Select data set
Mammary cancer
Programming references:
For the breast cancer data set
Data set partition method
Answer to others
Write your own code is mainly familiar with how to transfer packets, practice hands first. I wrote the code of breast cancer and found two problems: 1.10-fold CV can't compare with others [common division method] (https://www.bbsmax.com/A/QW5YW18Mzm/). 2. The accuracy estimated by loo is 0 ! [complex mood] (https://img-blog.csdnimg.cn/20200218154343443.jpg)
Here is the original code of breast cancer. Don't worry
#DATASET#1: breast cancer import numpy as np import pandas as pd from sklearn.metrics import classification_report #The following codes refer to https://www.bbsmax.com/A/QW5YW18Mzm/ # Create each column name columnNames = [ 'Sample code number', 'Clump Thickness', 'Uniformity of Cell Size', 'Uniformity of Cell Shape', 'Marginal Adhesion', 'Single Epithelial Cell Size', 'Bare Nuclei', 'Bland Chromatin', 'Normal Nucleoli', 'Mitoses', 'Class' ] data = pd.read_csv('http://archive.ics.uci.edu/ml/machine-learning-databases/breast-cancer-wisconsin/breast-cancer-wisconsin.data',names = columnNames)#If it's LOO, you need to supplement the parameter: delim_whitespace=True # Cleaning vacancy data data = data.replace(to_replace = "?", value = np.nan)#Replace lost data with "?" data = data.dropna(how = 'any')# And get rid of them X = data.iloc[:,0:10] Y = data.iloc[:,10] ##Refer to https://blog.csdn.net/snoopy yuan/article/details/64131129 for the following code #Regression of rate from sklearn.linear_model import LogisticRegression #metrics are evaluation modules, such as accuracy, etc from sklearn import metrics from sklearn.model_selection import cross_val_predict log_model=LogisticRegression() ''' #10fold CV, cross ﹣ val ﹣ predict returns the classification result of the estimator, which is used to compare with the actual data Y_pred = cross_val_predict(log_model,X,Y,cv=10) print("iris with 10folds, precision is:",metrics.accuracy_score(Y,Y_pred)) ''' ''' #--------------------------------Method split line------------------------------------------ #LOOCV from sklearn.model_selection import LeaveOneOut loo = LeaveOneOut() accuracy = 0#Because there is only one sample, the default is 0 #split is a method of leave one out model, which divides data into train and test arrays for train,test in loo.split(X): log_model.fit(X[train],Y[train]) #fit model Y_p=log_model.predict(X[test]) if Y_p==Y[test]: accuracy+=1 print("iris with LeaveOneOut, precision is:",accuracy/np.shape(X)[0]) #shape(x) is an array dimension, and shape(x)[0] is equal to the number of rows in the array, that is, the number of samples '''
Now check what's wrong ε = ('ο '))
Because the codes are all handled, and the big guys have no problems in experiments, they check them in blocks:
1. For blockා1, check the code of others, there are three differences:
- Normalization is not processed: other codes annotate the data normalization and then experiment, the results change little;
- Check whether it is related to the division method: the other code changes the test UU size to 0.1, and the result does not change much;
- The method of dividing feature matrix is different from that of labeling: the code of others is replaced by the following
X = data.iloc[:,0:10] Y = data.iloc[:,10] from sklearn.model_selection import train_test_split X_train, X_test, y_train, y_test = train_test_split( X, # features Y, # labels test_size = 0.1, random_state = 33 )
Output:
Accuracy of the LogesticRegression: 0.5217391304347826
OK, where is the wrong way to find it. Now change the code to:
Y_pred = cross_val_predict(log_model, data[ columnNames[1:10] ], # features data[ columnNames[10] ], # labels cv=10)
Despite a bunch of warnings, the output is
breast-cancer-wisconsin with 10folds, precision is: 0.9604685212298683
In some ways, it turned out to be almost the same. I gave so many warnings simply because I had 10% off and ran 10 times. This is the code to run:
# -*- coding: utf-8 -*- """ Created on Fri Feb 14 17:31:35 2020 @author: 29033 """ #DATASET#1: breast cancer import numpy as np import pandas as pd from sklearn.metrics import classification_report # Create each column name columnNames = [ 'Sample code number', 'Clump Thickness', 'Uniformity of Cell Size', 'Uniformity of Cell Shape', 'Marginal Adhesion', 'Single Epithelial Cell Size', 'Bare Nuclei', 'Bland Chromatin', 'Normal Nucleoli', 'Mitoses', 'Class' ] data = pd.read_csv('http://archive.ics.uci.edu/ml/machine-learning-databases/breast-cancer-wisconsin/breast-cancer-wisconsin.data',names = columnNames) # data processing data = data.replace(to_replace = "?", value = np.nan)#Replace lost data with "?" data = data.dropna(how = 'any')# And get rid of them X = data[ columnNames[1:10] ]# features Y = data[ columnNames[10] ]# labels #Regression of rate from sklearn.linear_model import LogisticRegression #metrics are evaluation modules, such as accuracy, etc from sklearn import metrics from sklearn.model_selection import cross_val_predict log_model=LogisticRegression() #10 fold cross validation Y_pred = cross_val_predict(log_model,X,Y,cv=10) print("breast-cancer-wisconsin with 10folds, precision is:",metrics.accuracy_score(Y,Y_pred))
Now focus on question 2. After modification, we ran and got the accuracy of 0.9633967789165446 [that is, there are more warnings]:
#--------------------------------Method split line------------------------------------------ #Leaving one method from sklearn.model_selection import LeaveOneOut loo = LeaveOneOut() accuracy = 0#Because there is only one sample, the default is 0 #split is a method of leave one out model, which divides data into train and test arrays for train,test in loo.split(X):#The type of loo.split(X) is < class' generator '> #There are 682 train s per time, 683 times, and the type is < class' numpy. Ndarray '> log_model.fit(X.iloc[train], Y.iloc[train]) # fitting Y_p = log_model.predict(X.iloc[test]) if (Y_p == Y.iloc[test]).any() : accuracy += 1 print("For the LOOCV, precision is:",accuracy/np.shape(X)[0]) #shape(x) is an array dimension, and shape(x)[0] is equal to the number of rows in the array, that is, the number of samples
Looking back, we found that the slice was wrong. X1 is the previous wrong slicing method. The comparison is as follows: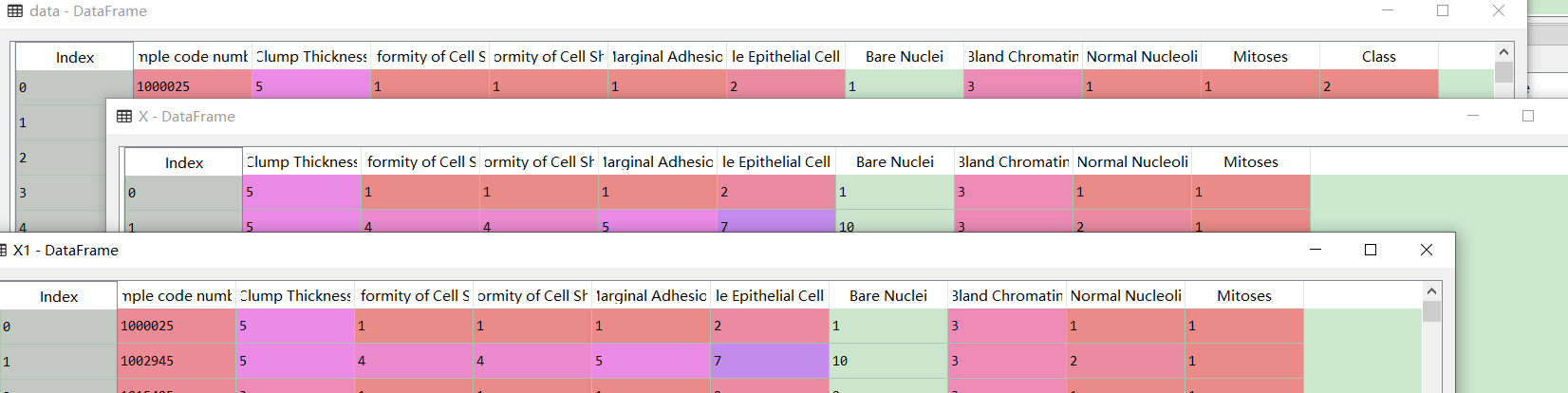 It should be changed to
It should be changed to
X2 = data.iloc[:,1:10] Y2 = data.iloc[:,10]
And label are responding, they are still symmetric after partition, no problem
After being beaten by life, select the data set iris, although there are Ready-made But it is recommended that you write your own code to enhance your proficiency.

