Summary:
SLIC: simple linear iterative clustering, a simple linear iterative clustering, uses k-means clustering to generate super-pixels, which is simple to implement, efficient to run, and has a better boundary preservation effect. The specific results compared with other methods are detailed in the following paper.
Paper address:https://infoscience.epfl.ch/record/177415/files/Superpixel_PAMI2011-2.pdf
1. Principles
SLIC essentially requires only one cluster number k, which is also the number of super pixels. The number of cluster centers is as many as the number of super pixels you want to divide the picture into. When initializing, the cluster centers are placed on the image at equal intervals. When the number of pixels in the image is set to N, the length of each edge is as follows:
N
/
k
\sqrt{N/k}
N/k
Place an initial cluster center in the square. To avoid using noise pixels and edges as cluster centers, move the center to the place with the smallest gradient in the surrounding 3*3 neighborhood.
Clustering with k-means is characterized by three values in the Lab color space plus coordinates x,y, a total of five dimensions.
x
i
.
f
e
a
t
u
r
e
=
[
L
,
a
,
b
,
x
,
y
]
T
x_{i.feature} = [L,a,b,x,y]^T
xi.feature =[L,a,b,x,y]T, and another difference from the regular k-means is that distance is calculated here only around the cluster center
2
N
/
k
∗
2
N
/
k
2\sqrt{N/k} * 2\sqrt{N/k}
2N/k
∗2N/k
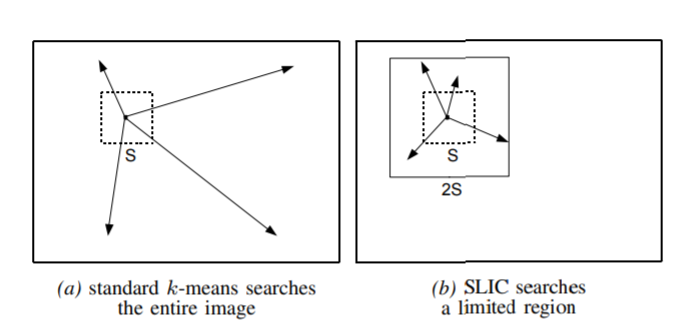
Because of its neighborhood, the computational effort is much smaller than that of the general k-mean clustering.Search space comparisons are shown below.
S
=
N
/
k
S = \sqrt{N/k}
S=N/k
After clustering, the average characteristics of each class are calculated, and the features of the cluster centers are updated to the average features.If the iteration is more than 10 times or the difference between before and after two clusters is less than the threshold value, then the end, otherwise continue clustering, update cluster centers...The algorithm flow is as follows:
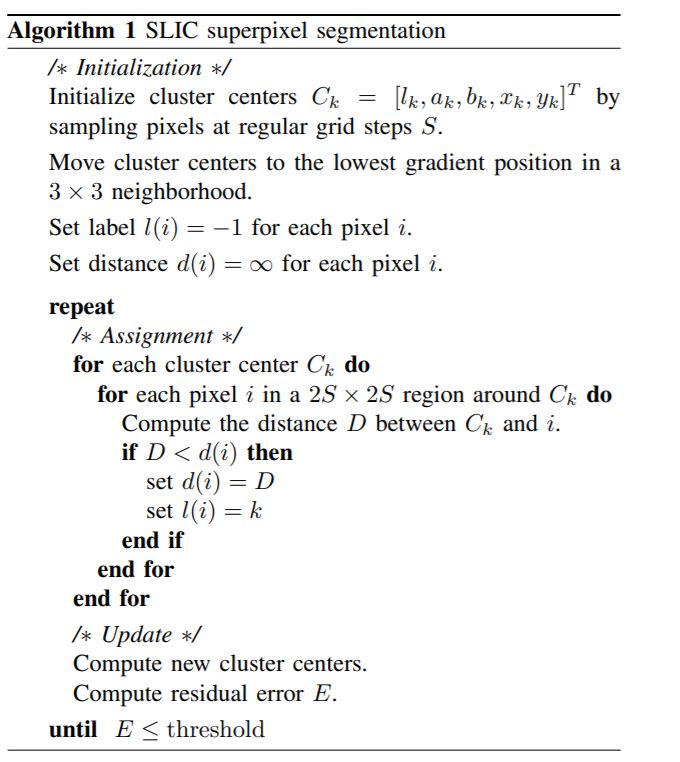
2. Implementation
2.1 Initializing Cluster Centers
In the original text, the cluster centers are initialized at equal intervals according to the number of clusters K. Assuming the total number of samples is N and the number of clusters is k, every other time
N
/
k
N/k
N/k samples are placed in a cluster center.Place K initial cluster centers on the image at equal intervals, that is, divide the image equally into equal edge lengths of
N
/
k
\sqrt{N/k}
N/k
Place the initial cluster center in a fixed position on the grid.In addition, to avoid the initial cluster center falling on the edge of the object, each cluster center has to be fine-tuned by calculating the gradient of the neighborhood around the initial cluster center point and moving the center point to the point with the smallest gradient.
It is intuitive to use the length len of the initial superpixel as a parameter to control the size of the generated superpixel. The relationship between k and Len is
l
e
n
=
N
/
k
len=\sqrt{N/k}
len=N/k
.
Note: Picture coordinate systems are originated in the upper left corner, positively oriented in the x-axis in the horizontal direction and positively oriented in the y-axis in the horizontal down direction (consistent with opencv). When accessing picture data matrices, they are usually described as rows and columns, that is, rows correspond to y and colums to X.
- Place cluster centers at equal intervals.
int initilizeCenters(cv::Mat &imageLAB, std::vector<center> ¢ers, int len)
{
if (imageLAB.empty())
{
std::cout << "In itilizeCenters: image is empty!\n";
return -1;
}
uchar *ptr = NULL;
center cent;
int num = 0;
for (int i = 0; i < imageLAB.rows; i += len)
{
cent.y = i + len / 2;
if (cent.y >= imageLAB.rows) continue;
ptr = imageLAB.ptr<uchar>(cent.y);
for (int j = 0; j < imageLAB.cols; j += len)
{
cent.x = j + len / 2;
if ((cent.x >= imageLAB.cols)) continue;
cent.L = *(ptr + cent.x * 3);
cent.A = *(ptr + cent.x * 3 + 1);
cent.B = *(ptr + cent.x * 3 + 2);
cent.label = ++num;
centers.push_back(cent);
}
}
return 0;
}
- Move the cluster center to the place with the smallest gradient in the surrounding 8 neighborhoods, and calculate the gradient using Sobel (you don't want the cluster center to fall on the edge to adjust it)
int fituneCenter(cv::Mat &imageLAB, cv::Mat &sobelGradient, std::vector<center> ¢ers)
{
if (sobelGradient.empty()) return -1;
center cent;
double *sobPtr = sobelGradient.ptr<double>(0);
uchar *imgPtr = imageLAB.ptr<uchar>(0);
int w = sobelGradient.cols;
for (int ck = 0; ck < centers.size(); ck++)
{
cent = centers[ck];
if (cent.x - 1 < 0 || cent.x + 1 >= sobelGradient.cols || cent.y - 1 < 0 || cent.y + 1 >= sobelGradient.rows)
{
continue;
}//end if
double minGradient = 9999999;
int tempx = 0, tempy = 0;
for (int m = -1; m < 2; m++)
{
sobPtr = sobelGradient.ptr<double>(cent.y + m);
for (int n = -1; n < 2; n++)
{
double gradient = pow(*(sobPtr + (cent.x + n) * 3), 2)
+ pow(*(sobPtr + (cent.x + n) * 3 + 1), 2)
+ pow(*(sobPtr + (cent.x + n) * 3 + 2), 2);
if (gradient < minGradient)
{
minGradient = gradient;
tempy = m;//row
tempx = n;//column
}//end if
}
}
cent.x += tempx;
cent.y += tempy;
imgPtr = imageLAB.ptr<uchar>(cent.y);
centers[ck].x = cent.x;
centers[ck].y = cent.y;
centers[ck].L = *(imgPtr + cent.x * 3);
centers[ck].A = *(imgPtr + cent.x * 3 + 1);
centers[ck].B = *(imgPtr + cent.x * 3 + 2);
}//end for
return 0;
}
2.2 Clustering
For each cluster center_k, calculates the distance from it to all points in the surrounding 2len*2len region, and if the newly computed distance is smaller than the original distance, the point is placed in center_k.
Notice that the nature of clustering is "objects are clustered". To judge the degree of "approximation" between sample points and cluster centers, one is distance measure: the closer the cluster is, the more similar the cluster centers are. The other is similarity measure, such as angular similarity coefficient, correlation coefficient, exponential similarity coefficient, etc.
There are many ways to calculate distance, such as:
- Euler Distance d ( x ⃗ , y ⃗ ) = ∣ ∣ x ⃗ − y ⃗ ∣ ∣ 2 2 d(\vec{x},\vec{y}) = ||\vec{x}-\vec{y}||_2^2 d(x ,y )=∣∣x −y ∣∣22
- Urban Distance d ( x ⃗ , y ⃗ ) = ∣ x ⃗ − y ⃗ ∣ d(\vec{x},\vec{y}) = |\vec{x}-\vec{y}| d(x ,y )=∣x −y ∣
- Chebyshev Distance d ( x ⃗ , y ⃗ ) = m a x ∣ x i − y i ∣ , i surface show dimension degree d(\vec{x},\vec{y}) = max|x_i-y_i|,i represents dimension d(x ,y )=max_xi_yi, i for dimension
- Mahalanobis: d ( x ⃗ , y ⃗ ) = ( x ⃗ − y ⃗ ) T V − 1 ( x ⃗ − y ⃗ ) , V surface show kind book total body Of Consortium square difference Moment front , V = 1 n − 1 Σ i = 1 n ( x i − x ⃗ ˉ ⃗ ) ( x i − x ⃗ ˉ ⃗ ) T , n by kind book number , x i ⃗ by No. i individual kind book ( column towards amount ) , x ⃗ ˉ by kind book all value D (\vec{x}, \vec{y}) = (\vec{x}-\vec{y})^TV^{1}(\vec{x}-\vec{y}), V represents the covariance matrix of the sample population, V= \frac{1}{n-1}\Sigma_{i=1}^{n}(\vec{x_i - \bar{\vec{x}} (\vec{x_i - \bar{\vec{x}}) ^T, n is the number of samples, \vec{x_i} is the first sample (column vector), \bar{\vec{x} is the sample mean d(x ,y )=(x −y )TV−1(x −y ), V represents the covariance matrix of the sample population, V=n_11Σi=1n (x I x) ˉ )(xi−x ˉ ) T,n is the number of samples, xi Is the f i rst sample (column vector), x ˉFor the sample mean, it has a good property that it is dimension independent (it also keeps the coordinate scale, rotation, translation unchanged, removes the correlation between components in a statistical sense), where the distance of the Lab color space is often much larger than the spatial distance when dividing the super-pixels.When Euclidean distance is used, a weight parameter is added to adjust the ratio of color distance to spatial distance.
If you have time in the future, consider trying out the effects of these distance clusters.The Euclidean distance is used here, and because the dimensions of Lab color space and image xy coordinate space are different, the weights of color space distance and xy coordinate distance need to be adjusted. The distance is calculated in the following way in this paper
d
c
=
(
l
j
−
l
i
)
2
+
(
a
j
−
a
i
)
2
+
(
b
j
−
b
i
)
2
d
s
=
(
x
j
−
x
i
)
2
+
(
y
j
−
y
i
)
2
D
=
d
c
2
+
(
d
s
S
)
2
m
2
\begin{aligned} d_{c}&=\sqrt{\left(l_{j}-l_{i}\right)^{2}+\left(a_{j}-a_{i}\right)^{2}+\left(b_{j}-b_{i}\right)^{2}} \\ d_{s} &=\sqrt{\left(x_{j}-x_{i}\right)^{2}+\left(y_{j}-y_{i}\right)^{2}}\\D&=\sqrt{d_{c}^{2}+\left(\frac{d_{s}}{S}\right)^{2} m^{2}} \end{aligned}
dcdsD=(lj−li)2+(aj−ai)2+(bj−bi)2
=(xj−xi)2+(yj−yi)2
=dc2+(Sds)2m2
But in fact, when we do super-pixel segmentation, we are more concerned about the size of the super-pixels than how many. Although there is a clear correspondence between the size S and the number of clusters k, the input parameter k is not as direct as that of size S. In addition
d
s
Of
power
heavy
use
m
2
S
2
Weight Use of ds\frac{m^2}{S^2}
The weight of ds is actually a little cumbersome with S2m2, because modifying m or s individually will be modulated by another parameter, so I changed the calculation of D to the following
D
=
d
c
2
+
m
d
s
2
\begin{aligned}D=\sqrt{d_{c}^{2}+md_{s}^{2} }\end{aligned}
D=dc2+mds2
int clustering(const cv::Mat &imageLAB, cv::Mat &DisMask, cv::Mat &labelMask,
std::vector<center> ¢ers, int len, int m)
{
if (imageLAB.empty())
{
std::cout << "clustering :the input image is empty!\n";
return -1;
}
double *disPtr = NULL;//disMask type: 64FC1
double *labelPtr = NULL;//labelMask type: 64FC1
const uchar *imgPtr = NULL;//imageLAB type: 8UC3
//disc = std::sqrt(pow(L - cL, 2)+pow(A - cA, 2)+pow(B - cB,2))
//diss = std::sqrt(pow(x-cx,2) + pow(y-cy,2));
//dis = sqrt(disc^2 + (diss/len)^2 * m^2)
double dis = 0, disc = 0, diss = 0;
//cluster center's cx, cy,cL,cA,cB;
int cx, cy, cL, cA, cB, clabel;
//imageLAB's x, y, L,A,B
int x, y, L, A, B;
//Note: The image coordinates here are from the upper left corner, x-direction horizontally to the right, y-direction horizontally downward, consistent with opencv
// From the matrix row and column perspective, I represents the row and j represents the column, i.e. (i,j) = (y,x)
for (int ck = 0; ck < centers.size(); ++ck)
{
cx = centers[ck].x;
cy = centers[ck].y;
cL = centers[ck].L;
cA = centers[ck].A;
cB = centers[ck].B;
clabel = centers[ck].label;
for (int i = cy - len; i < cy + len; i++)
{
if (i < 0 | i >= imageLAB.rows) continue;
//pointer point to the ith row
imgPtr = imageLAB.ptr<uchar>(i);
disPtr = DisMask.ptr<double>(i);
labelPtr = labelMask.ptr<double>(i);
for (int j = cx - len; j < cx + len; j++)
{
if (j < 0 | j >= imageLAB.cols) continue;
L = *(imgPtr + j * 3);
A = *(imgPtr + j * 3 + 1);
B = *(imgPtr + j * 3 + 2);
disc = std::sqrt(pow(L - cL, 2) + pow(A - cA, 2) + pow(B - cB, 2));
diss = std::sqrt(pow(j - cx, 2) + pow(i - cy, 2));
dis = sqrt(pow(disc, 2) + m * pow(diss, 2));
if (dis < *(disPtr + j))
{
*(disPtr + j) = dis;
*(labelPtr + j) = clabel;
}//end if
}//end for
}
}//end for (int ck = 0; ck < centers.size(); ++ck)
return 0;
}
2.3 Update Cluster Center
For each cluster center_k, averaging the features of all points belonging to this class, assigning the average to center_k.
int updateCenter(cv::Mat &imageLAB, cv::Mat &labelMask, std::vector<center> ¢ers, int len)
{
double *labelPtr = NULL;//labelMask type: 64FC1
const uchar *imgPtr = NULL;//imageLAB type: 8UC3
int cx, cy;
for (int ck = 0; ck < centers.size(); ++ck)
{
double sumx = 0, sumy = 0, sumL = 0, sumA = 0, sumB = 0, sumNum = 0;
cx = centers[ck].x;
cy = centers[ck].y;
for (int i = cy - len; i < cy + len; i++)
{
if (i < 0 | i >= imageLAB.rows) continue;
//pointer point to the ith row
imgPtr = imageLAB.ptr<uchar>(i);
labelPtr = labelMask.ptr<double>(i);
for (int j = cx - len; j < cx + len; j++)
{
if (j < 0 | j >= imageLAB.cols) continue;
if (*(labelPtr + j) == centers[ck].label)
{
sumL += *(imgPtr + j * 3);
sumA += *(imgPtr + j * 3 + 1);
sumB += *(imgPtr + j * 3 + 2);
sumx += j;
sumy += i;
sumNum += 1;
}//end if
}
}
//update center
if (sumNum == 0) sumNum = 0.000000001;
centers[ck].x = sumx / sumNum;
centers[ck].y = sumy / sumNum;
centers[ck].L = sumL / sumNum;
centers[ck].A = sumA / sumNum;
centers[ck].B = sumB / sumNum;
}//end for
return 0;
}
2.4 Display super-pixel segmentation results
Mode 1: Replace the features of points belonging to the same class with average features;
Mode 2: Draw the cluster boundary;
3. Measured results
- The left side is the original, the middle part is the super-pixel boundary effect, and the right side is the super-pixel image effect







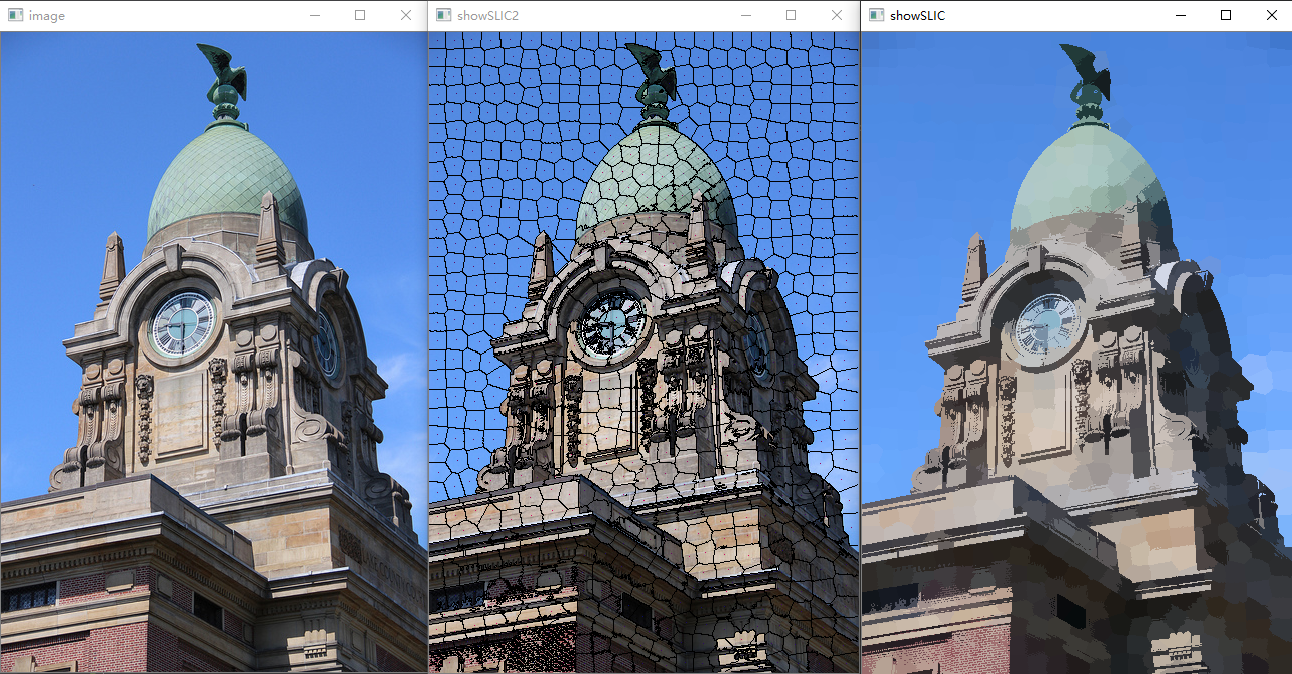
4. Full Source
//
//created by Mr. Peng. 2021\08\31
//
#include "opencv.hpp"
struct center
{
int x;//column
int y;//row
int L;
int A;
int B;
int label;
};
/
//input parameters:
//imageLAB: the source image in Lab color space
//DisMask: it save the shortest distance to the nearest center
//labelMask: it save every pixel's label
//centers: clustering center
//len: the super pixls will be initialize to len*len
//m: a parameter witch adjust the weights of the spacial and color space distance
//
//output:
int clustering(const cv::Mat &imageLAB, cv::Mat &DisMask, cv::Mat &labelMask,
std::vector<center> ¢ers, int len, int m)
{
if (imageLAB.empty())
{
std::cout << "clustering :the input image is empty!\n";
return -1;
}
double *disPtr = NULL;//disMask type: 64FC1
double *labelPtr = NULL;//labelMask type: 64FC1
const uchar *imgPtr = NULL;//imageLAB type: 8UC3
//disc = std::sqrt(pow(L - cL, 2)+pow(A - cA, 2)+pow(B - cB,2))
//diss = std::sqrt(pow(x-cx,2) + pow(y-cy,2));
//dis = sqrt(disc^2 + (diss/len)^2 * m^2)
double dis = 0, disc = 0, diss = 0;
//cluster center's cx, cy,cL,cA,cB;
int cx, cy, cL, cA, cB, clabel;
//imageLAB's x, y, L,A,B
int x, y, L, A, B;
//Note: The image coordinates here are from the upper left corner, x-direction horizontally to the right, y-direction horizontally downward, consistent with opencv
// From the matrix row and column perspective, I represents the row and j represents the column, i.e. (i,j) = (y,x)
for (int ck = 0; ck < centers.size(); ++ck)
{
cx = centers[ck].x;
cy = centers[ck].y;
cL = centers[ck].L;
cA = centers[ck].A;
cB = centers[ck].B;
clabel = centers[ck].label;
for (int i = cy - len; i < cy + len; i++)
{
if (i < 0 | i >= imageLAB.rows) continue;
//pointer point to the ith row
imgPtr = imageLAB.ptr<uchar>(i);
disPtr = DisMask.ptr<double>(i);
labelPtr = labelMask.ptr<double>(i);
for (int j = cx - len; j < cx + len; j++)
{
if (j < 0 | j >= imageLAB.cols) continue;
L = *(imgPtr + j * 3);
A = *(imgPtr + j * 3 + 1);
B = *(imgPtr + j * 3 + 2);
disc = std::sqrt(pow(L - cL, 2) + pow(A - cA, 2) + pow(B - cB, 2));
diss = std::sqrt(pow(j - cx, 2) + pow(i - cy, 2));
dis = sqrt(pow(disc, 2) + m * pow(diss, 2));
if (dis < *(disPtr + j))
{
*(disPtr + j) = dis;
*(labelPtr + j) = clabel;
}//end if
}//end for
}
}//end for (int ck = 0; ck < centers.size(); ++ck)
return 0;
}
/
//input parameters:
//imageLAB: the source image in Lab color space
//labelMask: it save every pixel's label
//centers: clustering center
//len: the super pixls will be initialize to len*len
//
//output:
int updateCenter(cv::Mat &imageLAB, cv::Mat &labelMask, std::vector<center> ¢ers, int len)
{
double *labelPtr = NULL;//labelMask type: 64FC1
const uchar *imgPtr = NULL;//imageLAB type: 8UC3
int cx, cy;
for (int ck = 0; ck < centers.size(); ++ck)
{
double sumx = 0, sumy = 0, sumL = 0, sumA = 0, sumB = 0, sumNum = 0;
cx = centers[ck].x;
cy = centers[ck].y;
for (int i = cy - len; i < cy + len; i++)
{
if (i < 0 | i >= imageLAB.rows) continue;
//pointer point to the ith row
imgPtr = imageLAB.ptr<uchar>(i);
labelPtr = labelMask.ptr<double>(i);
for (int j = cx - len; j < cx + len; j++)
{
if (j < 0 | j >= imageLAB.cols) continue;
if (*(labelPtr + j) == centers[ck].label)
{
sumL += *(imgPtr + j * 3);
sumA += *(imgPtr + j * 3 + 1);
sumB += *(imgPtr + j * 3 + 2);
sumx += j;
sumy += i;
sumNum += 1;
}//end if
}
}
//update center
if (sumNum == 0) sumNum = 0.000000001;
centers[ck].x = sumx / sumNum;
centers[ck].y = sumy / sumNum;
centers[ck].L = sumL / sumNum;
centers[ck].A = sumA / sumNum;
centers[ck].B = sumB / sumNum;
}//end for
return 0;
}
int showSLICResult(const cv::Mat &image, cv::Mat &labelMask, std::vector<center> ¢ers, int len)
{
cv::Mat dst = image.clone();
cv::cvtColor(dst, dst, cv::COLOR_BGR2Lab);
double *labelPtr = NULL;//labelMask type: 32FC1
uchar *imgPtr = NULL;//image type: 8UC3
int cx, cy;
double sumx = 0, sumy = 0, sumL = 0, sumA = 0, sumB = 0, sumNum = 0.00000001;
for (int ck = 0; ck < centers.size(); ++ck)
{
cx = centers[ck].x;
cy = centers[ck].y;
for (int i = cy - len; i < cy + len; i++)
{
if (i < 0 | i >= image.rows) continue;
//pointer point to the ith row
imgPtr = dst.ptr<uchar>(i);
labelPtr = labelMask.ptr<double>(i);
for (int j = cx - len; j < cx + len; j++)
{
if (j < 0 | j >= image.cols) continue;
if (*(labelPtr + j) == centers[ck].label)
{
*(imgPtr + j * 3) = centers[ck].L;
*(imgPtr + j * 3 + 1) = centers[ck].A;
*(imgPtr + j * 3 + 2) = centers[ck].B;
}//end if
}
}
}//end for
cv::cvtColor(dst, dst, cv::COLOR_Lab2BGR);
cv::namedWindow("showSLIC", 0);
cv::imshow("showSLIC", dst);
cv::waitKey(1);
return 0;
}
int showSLICResult2(const cv::Mat &image, cv::Mat &labelMask, std::vector<center> ¢ers, int len)
{
cv::Mat dst = image.clone();
//cv::cvtColor(dst, dst, cv::COLOR_Lab2BGR);
double *labelPtr = NULL;//labelMask type: 32FC1
double *labelPtr_nextRow = NULL;//labelMask type: 32FC1
uchar *imgPtr = NULL;//image type: 8UC3
for (int i = 0; i < labelMask.rows - 1; i++)
{
labelPtr = labelMask.ptr<double>(i);
imgPtr = dst.ptr<uchar>(i);
for (int j = 0; j < labelMask.cols - 1; j++)
{
//if left pixel's label is different from the right's
if (*(labelPtr + j) != *(labelPtr + j + 1))
{
*(imgPtr + 3 * j) = 0;
*(imgPtr + 3 * j + 1) = 0;
*(imgPtr + 3 * j + 2) = 0;
}
//if the upper pixel's label is different from the bottom's
labelPtr_nextRow = labelMask.ptr<double>(i + 1);
if (*(labelPtr_nextRow + j) != *(labelPtr + j))
{
*(imgPtr + 3 * j) = 0;
*(imgPtr + 3 * j + 1) = 0;
*(imgPtr + 3 * j + 2) = 0;
}
}
}
//show center
for (int ck = 0; ck < centers.size(); ck++)
{
imgPtr = dst.ptr<uchar>(centers[ck].y);
*(imgPtr + centers[ck].x * 3) = 100;
*(imgPtr + centers[ck].x * 3 + 1) = 100;
*(imgPtr + centers[ck].x * 3 + 1) = 10;
}
cv::namedWindow("showSLIC2", 0);
cv::imshow("showSLIC2", dst);
cv::waitKey(1);
return 0;
}
int initilizeCenters(cv::Mat &imageLAB, std::vector<center> ¢ers, int len)
{
if (imageLAB.empty())
{
std::cout << "In itilizeCenters: image is empty!\n";
return -1;
}
uchar *ptr = NULL;
center cent;
int num = 0;
for (int i = 0; i < imageLAB.rows; i += len)
{
cent.y = i + len / 2;
if (cent.y >= imageLAB.rows) continue;
ptr = imageLAB.ptr<uchar>(cent.y);
for (int j = 0; j < imageLAB.cols; j += len)
{
cent.x = j + len / 2;
if ((cent.x >= imageLAB.cols)) continue;
cent.L = *(ptr + cent.x * 3);
cent.A = *(ptr + cent.x * 3 + 1);
cent.B = *(ptr + cent.x * 3 + 2);
cent.label = ++num;
centers.push_back(cent);
}
}
return 0;
}
//if the center locates in the edges, fitune it's location.
int fituneCenter(cv::Mat &imageLAB, cv::Mat &sobelGradient, std::vector<center> ¢ers)
{
if (sobelGradient.empty()) return -1;
center cent;
double *sobPtr = sobelGradient.ptr<double>(0);
uchar *imgPtr = imageLAB.ptr<uchar>(0);
int w = sobelGradient.cols;
for (int ck = 0; ck < centers.size(); ck++)
{
cent = centers[ck];
if (cent.x - 1 < 0 || cent.x + 1 >= sobelGradient.cols || cent.y - 1 < 0 || cent.y + 1 >= sobelGradient.rows)
{
continue;
}//end if
double minGradient = 9999999;
int tempx = 0, tempy = 0;
for (int m = -1; m < 2; m++)
{
sobPtr = sobelGradient.ptr<double>(cent.y + m);
for (int n = -1; n < 2; n++)
{
double gradient = pow(*(sobPtr + (cent.x + n) * 3), 2)
+ pow(*(sobPtr + (cent.x + n) * 3 + 1), 2)
+ pow(*(sobPtr + (cent.x + n) * 3 + 2), 2);
if (gradient < minGradient)
{
minGradient = gradient;
tempy = m;//row
tempx = n;//column
}//end if
}
}
cent.x += tempx;
cent.y += tempy;
imgPtr = imageLAB.ptr<uchar>(cent.y);
centers[ck].x = cent.x;
centers[ck].y = cent.y;
centers[ck].L = *(imgPtr + cent.x * 3);
centers[ck].A = *(imgPtr + cent.x * 3 + 1);
centers[ck].B = *(imgPtr + cent.x * 3 + 2);
}//end for
return 0;
}
/
//input parameters:
//image: the source image in RGB color space
//resultLabel: it save every pixel's label
//len: the super pixls will be initialize to len*len
//m: a parameter witch adjust the weights of diss
//output:
int SLIC(cv::Mat &image, cv::Mat &resultLabel, std::vector<center> ¢ers, int len, int m)
{
if (image.empty())
{
std::cout << "in SLIC the input image is empty!\n";
return -1;
}
int MAXDIS = 999999;
int height, width;
height = image.rows;
width = image.cols;
//convert color
cv::Mat imageLAB;
cv::cvtColor(image, imageLAB, cv::COLOR_BGR2Lab);
//get sobel gradient map
cv::Mat sobelImagex, sobelImagey, sobelGradient;
cv::Sobel(imageLAB, sobelImagex, CV_64F, 0, 1, 3);
cv::Sobel(imageLAB, sobelImagey, CV_64F, 1, 0, 3);
cv::addWeighted(sobelImagex, 0.5, sobelImagey, 0.5, 0, sobelGradient);//sobel output image type is CV_64F
//initiate
//std::vector<center> centers;
//disMask save the distance of the pixels to center;
cv::Mat disMask ;
//labelMask save the label of the pixels
cv::Mat labelMask = cv::Mat::zeros(cv::Size(width, height), CV_64FC1);
//initialize centers, get centers
initilizeCenters(imageLAB, centers, len);
//if the center locates in the edges, fitune it's location
fituneCenter(imageLAB, sobelGradient, centers);
//update cluster 10 times
for (int time = 0; time < 10; time++)
{
//clustering
disMask = cv::Mat(height, width, CV_64FC1, cv::Scalar(MAXDIS));
clustering(imageLAB, disMask, labelMask, centers, len, m);
//update
updateCenter(imageLAB, labelMask, centers, len);
//fituneCenter(imageLAB, sobelGradient, centers);
}
resultLabel = labelMask;
return 0;
}
int SLIC_Demo()
{
std::string imagePath = "K:\\deepImage\\plane.jpg";
cv::Mat image = cv::imread(imagePath);
cv::Mat labelMask;//save every pixel's label
cv::Mat dst;//save the shortest distance to the nearest centers
std::vector<center> centers;//clustering centers
int len = 25;//the scale of the superpixel ,len*len
int m = 10;//a parameter witch adjust the weights of spacial distance and the color space distance
SLIC(image, labelMask, centers, len, m);
cv::namedWindow("image", 1);
cv::imshow("image", image);
showSLICResult(image, labelMask, centers, len);
showSLICResult2(image, labelMask, centers, len);
cv::waitKey(0);
return 0;
}
int main()
{
SLIC_Demo();
return 0;
}