Python data analysis and presentation
0.conda and IPython
anaconda=conda + Python of a certain version + third-party library
conda: package and environment management tools
IPython
?
Add a before or after the variable? Display general information
%Command
| command | meaning |
|---|---|
| %run | Execute% (execute external code) in an empty namespace |
| %magic | Show all magic commands |
| %hist | IPython command input history |
| %pdb | Automatically enter the debugger after an exception occurs |
| %reset | Empty namespace |
| %who | Displays the variables defined in the namespace |
| %time statement | Give code execution time |
| %timeit statement | Execute the code multiple times and calculate the comprehensive average execution time |
| %paste/%cpaste | Don't mess the format when copying code |
| %timeit | Calculate code run time |
1.Numpy Library
Difference between list and array
The list data types can be different, and the array data types are the same
N-dimensional array object: ndarray
-
The bottom layer of numpy is implemented by C
-
np.array() generates an array of ndarray s
-
axis: the dimension to save data
-
rank: number of dimensions (number of axes)
ndarray object properties
| attribute | explain |
|---|---|
| .ndim | Number of dimensions (number of axes) |
| .shape | The scale of ndarray is n rows and m columns for a matrix |
| .size | Number of ndarray elements, equivalent to n*m in. shape |
| .dtype | ndarray element type |
| .itemsize | The size of the ndarray element in bytes |
ndarray element type
| data type | explain |
|---|---|
| bool | True or False |
| intc | Same as int (32 or 64) in C language |
| intp | Integer used for index, the same as ssize in C_ T (32 or 64) |
| int8 | 8-bit int |
| int16 | 16 bit int |
| int32 | 32-bit int |
| int64 | 64 bit int |
| uint8 | 8-bit unsigned |
| uint16 | 16 bit unsigned |
| uint32 | 32-bit unsigned |
| uint64 | 64 bit unsigned |
| float16 | 1+5+10 |
| float32 | 1+8+23 |
| float64 | 1+11+52 |
| complex64 | The complex number, real part and imaginary part are 32-bit floating-point numbers |
| complex128 | The complex number, real part and imaginary part are 64 bit floating-point numbers |
Create method
(1) Create ndarray from list, tuple
#list
>>> x=np.array([0,1,2,3])
>>> x
array([0, 1, 2, 3])
#tuple
>>> x=np.array((1,2,3))
>>> x
array([1, 2, 3])
#List tuple mixing
>>> x=np.array([[1,2],[9,8],(0.1,0.2)])
>>> x
array([[1. , 2. ],
[9. , 8. ],
[0.1, 0.2]])
(2) Functions provided by Numpy
| function | explain |
|---|---|
| np.arange(n) | Returns ndarray, from 0 to n-1 |
| np.ones(shape) | Generate a full 1 array, and shape is a tuple |
| np.zeros(shape) | Generate all 0 arrays, and shape is a tuple |
| np.full(shape,val) | Generate a full val array. shape is a tuple |
| np.eye(n) | Generate n*n diagonal matrix |
| np.ones_like(a) | Generate a full 1 array according to the shape of A |
| np.zeros_like(a) | Generate an array of all zeros according to the shape of a |
| np.full_like(a,val) | Generate a full val array according to the shape of A |
(3) Other functions
| function | explain |
|---|---|
| np.linspace() | Fill the data evenly according to the start and end data to form an array |
| np.concatenate() | Merge two or more arrays |
example:
>>> a=np.linspace(1,10,4) >>> a array([ 1., 4., 7., 10.]) >>> b=np.linspace(1,10,4,endpoint=False) >>> b array([1. , 3.25, 5.5 , 7.75]) >>> c=np.concatenate((a,b)) >>> c array([ 1. , 4. , 7. , 10. , 1. , 3.25, 5.5 , 7.75])
Dimension transformation
| method | explain |
|---|---|
| .reshape(shape) | Without changing the original array, an array of shape shapes is returned according to the original array |
| .resize(shape) | Transform the original array into shape |
| .swapaxes(ax1,ax2) | Swap two of the n dimensions in the array |
| .flatten() | Returns the folded one-dimensional array without changing the original array |
Array type transformation
>>> a=np.ones((2,3,4))
>>> a
array([[[1., 1., 1., 1.],
[1., 1., 1., 1.],
[1., 1., 1., 1.]],
[[1., 1., 1., 1.],
[1., 1., 1., 1.],
[1., 1., 1., 1.]]])
>>> a=np.ones((2,3,4),dtype=np.int64)
>>> a
array([[[1, 1, 1, 1],
[1, 1, 1, 1],
[1, 1, 1, 1]],
[[1, 1, 1, 1],
[1, 1, 1, 1],
[1, 1, 1, 1]]], dtype=int64)
>>> b=a.astype(np.float64)
>>> b
array([[[1., 1., 1., 1.],
[1., 1., 1., 1.],
[1., 1., 1., 1.]],
[[1., 1., 1., 1.],
[1., 1., 1., 1.],
[1., 1., 1., 1.]]])
Array - > List
>>> a=np.full((2,3,4),25,dtype=np.int32)
>>> a
array([[[25, 25, 25, 25],
[25, 25, 25, 25],
[25, 25, 25, 25]],
[[25, 25, 25, 25],
[25, 25, 25, 25],
[25, 25, 25, 25]]])
>>> a.tolist()
[[[25, 25, 25, 25], [25, 25, 25, 25], [25, 25, 25, 25]], [[25, 25, 25, 25], [25, 25, 25, 25], [25, 25, 25, 25]]]
Array index and slice
One dimensional array
>>> a=np.array([9,8,7,6,5]) >>> a[2] 7 >>> a[1:4:2] array([8, 6])
Multidimensional array
>>> a=np.array([9,8,7,6,5])
>>> a[2]
7
>>> a[1:4:2]
array([8, 6])
>>> a=np.arange(24).reshape((2,3,4))
>>> a[1,2,3]
23
>>> a[:,1,-3]#Do not care about the first dimension. The second dimension is required to be 1 and the third dimension is - 3
array([ 5, 17])
>>> a[:,:,::2]#Do not care about the first and second dimensions, and the step size of the third dimension is 2
array([[[ 0, 2],
[ 4, 6],
[ 8, 10]],
[[12, 14],
[16, 18],
[20, 22]]])
Array operation
>>> a=np.arange(24).reshape((2,3,4))
>>> a
array([[[ 0, 1, 2, 3],
[ 4, 5, 6, 7],
[ 8, 9, 10, 11]],
[[12, 13, 14, 15],
[16, 17, 18, 19],
[20, 21, 22, 23]]])
>>> a=a/a.mean()
>>> a
array([[[0. , 0.08695652, 0.17391304, 0.26086957],
[0.34782609, 0.43478261, 0.52173913, 0.60869565],
[0.69565217, 0.7826087 , 0.86956522, 0.95652174]],
[[1.04347826, 1.13043478, 1.2173913 , 1.30434783],
[1.39130435, 1.47826087, 1.56521739, 1.65217391],
[1.73913043, 1.82608696, 1.91304348, 2. ]]])
| function | explain |
|---|---|
| np.abs(x) np.fabs(x) | Calculate the absolute value of each element of the array |
| np.sqrt(x) | Calculate the square root of each element of the array |
| np.square(x) | Calculate the square of each element of the array |
| np.log(x) np.log10(x) np.log2(x) | Calculate the natural logarithm, base 10 logarithm and base 2 logarithm of each element of the array |
| np.ceil(x) np.floor(x) | Calculate the ceiling value and floor value of each element of the array |
| np.rint(x) | Calculates the rounded value of each element of the array |
| np.modf(x) | Returns the decimal and integer parts of each element of the array as two independent arrays |
| np.cos(x) np.cosh(x) and so on | Calculate the trigonometric function and hyperbolic function of each element |
| np.exp(x) | Calculate the index value of each element of the array |
| np.sign(x) | Calculate the symbolic value of each element of the array 10 |
| + - * / ** | Each element of the two arrays performs corresponding operation |
| np.maximum(x,y) np.fmax() | Calculation of maximum and minimum values at element level |
| np.copysign(x,y) | Assign the sign of each element value in array y to the corresponding element of array x |
| np.mod(x,y) | Modular operation at element level |
| > < >= <= == != | Arithmetic comparison to produce Boolean arrays |
>>> import numpy as np
>>> a=np.arange(24).reshape((2,3,4))
>>> np.square(a)
array([[[ 0, 1, 4, 9],
[ 16, 25, 36, 49],
[ 64, 81, 100, 121]],
[[144, 169, 196, 225],
[256, 289, 324, 361],
[400, 441, 484, 529]]], dtype=int32)
>>> a=np.sqrt(a)
>>> a
array([[[0. , 1. , 1.41421356, 1.73205081],
[2. , 2.23606798, 2.44948974, 2.64575131],
[2.82842712, 3. , 3.16227766, 3.31662479]],
[[3.464 one hundred and one 62, 3.60555128, 3.74165739, 3.87298335],
[4. , 4.12310563, 4.24264069, 4.35889894],
[4.47213595, 4.58257569, 4.69041576, 4.79583152]]])
>>> np.modf(a)
(array([[[0. , 0. , 0.41421356, 0.73205081],
[0. , 0.23606798, 0.44948974, 0.64575131],
[0.82842712, 0. , 0.16227766, 0.31662479]],
[[0.46410162, 0.60555128, 0.74165739, 0.87298335],
[0. , 0.12310563, 0.24264069, 0.35889894],
[0.47213595, 0.58257569, 0.69041576, 0.79583152]]]), array([[[0., 1., 1., 1.],
[2., 2., 2., 2.],
[2., 3., 3., 3.]],
[[3., 3., 3., 3.],
[4., 4., 4., 4.],
[4., 4., 4., 4.]]]))
>>> a=np.arange(24).reshape((2,3,4))
>>> b=np.sqrt(a)
>>> a
array([[[ 0, 1, 2, 3],
[ 4, 5, 6, 7],
[ 8, 9, 10, 11]],
[[12, 13, 14, 15],
[16, 17, 18, 19],
[20, 21, 22, 23]]])
>>> b
array([[[0. , 1. , 1.41421356, 1.73205081],
[2. , 2.23606798, 2.44948974, 2.64575131],
[2.82842712, 3. , 3.16227766, 3.31662479]],
[[3.46410162, 3.60555128, 3.74165739, 3.87298335],
[4. , 4.12310563, 4.24264069, 4.35889894],
[4.47213595, 4.58257569, 4.69041576, 4.79583152]]])
>>> np.maximum(a,b)
array([[[ 0., 1., 2., 3.],
[ 4., 5., 6., 7.],
[ 8., 9., 10., 11.]],
[[12., 13., 14., 15.],
[16., 17., 18., 19.],
[20., 21., 22., 23.]]])
>>> a>b
array([[[False, False, True, True],
[ True, True, True, True],
[ True, True, True, True]],
[[ True, True, True, True],
[ True, True, True, True],
[ True, True, True, True]]])
Write csv
np.savetxt(fname,array,fmt='%.18e',delimiter=None)
- frame: file, string or generator, which can be a compressed file of. gz or. bz2
- Array: the array stored in the file
- fmt: the format in which the file is written, e.g.% d,%.2f,%.18e
- delimiter: splits a string. The default is a space
a=np.array(100).reshape(5,20)
np.savetxt('a.csv',a,fmt='%d',delimiter=',')
0,1,2,3...99
a=np.array(100).reshape(5,20)
np.savetxt('a.csv',a,fmt='%1f',delimiter=',')
0.0,1.0,2.0...99.0
Read csv
np.loadtxt(fname,dtype=np.float,delimiter=None,unpack=False)
- frame: file, string or generator, which can be a compressed file of. gz or. bz2
- dtype: data type; optional
- delimiter: split string, default space
- unpack: if True, the read in properties will be written to different variables respectively
b=np.loadtxt('a.csv',delimiter=',')
b=np.loadtxt('a.csv',dtype=int,delimiter=',')
Multidimensional data access
The above savetxt and loadtext can only handle one-dimensional and two-dimensional arrays
a.tofile(fname,sep='',format='%s')
- frame: file, string
- sep: data split string. If it is an empty string, the written file is binary
- Format: the format of the written data
a=np.arange(100).reshape(5,10,2)
a.tofile('b.dat',sep=',',format='%d')
#Generating binaries without specifying sep
np.fromfile(fname,dtype=float,count=-1,sep='')
- frame: file, string
- dtype: read data type
- count: the number of elements read in, - 1 means the whole file is read in
- sep: data split string. If it is an empty string, the written file is binary
a=np.arange(100).reshape(5,10,2)
a.tofile('b.dat',sep=',',format='%d')
c=np.fromfile('b.dat',dtype=np.int,sep=",")
c=np.fromfile('b.dat',dtype=np.int,sep=",").reshape(5,10,2)
Convenient file access
np.save(fname,array) np.savez(fname,array) np.load(fname)
- fname: file name, with. npy as the extension and npz as the compression extension
- Array: array variable
a=np.arange(100).reshape(5,10,2)
np.save("a.npy",a)
b=np.load("a.npy")
Random number function
| function | explain |
|---|---|
| rand(d0,d1...dn) | Create a random number array according to d0 DN, floating point number [0,1], evenly distributed |
| randn(d0,d1...dn) | Create a random number array according to d0 DN, with standard normal distribution |
| randint(low[,high,shape]) | Create random integers or array of integers according to shape (the range is [low,high) |
| seed(s) | Random number seed, s is the given seed value |
>>> a=np.random.rand(3,4,5)
>>> a
array([[[0.31789129, 0.04067915, 0.83303674, 0.68403089, 0.8170608 ],
[0.48896229, 0.87662241, 0.80402316, 0.65002092, 0.60505475],
[0.15862944, 0.86727453, 0.52962166, 0.60726439, 0.82176069],
[0.12508611, 0.42132045, 0.31606682, 0.35658386, 0.19045285]],
[[0.89511544, 0.26119674, 0.64866715, 0.23401689, 0.95676794],
[0.43291969, 0.6155749 , 0.92665404, 0.05817955, 0.51062756],
[0.10594579, 0.73809733, 0.47707574, 0.07499143, 0.11235247],
[0.1250221 , 0.7002249 , 0.69170791, 0.74646681, 0.63357624]],
[[0.50299485, 0.67903174, 0.73188944, 0.6296878 , 0.98629917],
[0.8952625 , 0.76823821, 0.48124231, 0.02401954, 0.41473022],
[0.72505748, 0.31233367, 0.81575195, 0.19943229, 0.60471366],
[0.90742952, 0.89124879, 0.1213489 , 0.88284548, 0.67810859]]])
>>> sn=np.random.randn(3,4,5)
>>> sn
array([[[ 1.1832839 , 0.25060466, -0.57017814, 0.47517595,
-1.51740385],
[ 0.35038796, -1.82168871, 0.16061641, -0.88696559,
0.94106256],
[ 0.77097301, 1.11086675, 0.32795292, -0.9581694 ,
-1.75591931],
[-0.14176509, 1.68687842, 1.30323321, 0.30734734,
-0.42623733]],
[[ 1.00836129, -1.4763877 , -0.10079373, -0.631505 ,
-0.33870885],
[-0.88316216, -1.5023321 , -0.51582272, 0.46557721,
1.47694773],
[-0.4808448 , 0.88825249, -0.07169483, -0.03960847,
2.03167751],
[ 1.38474762, 0.86621196, 0.68230671, -0.75887396,
-0.41100159]],
[[-0.18006775, 0.50978947, -0.2241126 , -1.37232262,
0.74588361],
[-0.77222672, 0.06994181, -0.23934626, 0.43249514,
0.40141005],
[-1.35859946, 1.47103075, 0.59458256, -1.81122503,
0.93659709],
[-0.80001237, -0.46711391, 0.70415531, 0.2639113 ,
-0.17576713]]])
>>> b=np.random.randint(100,200,(3,4))
>>> b
array([[164, 145, 135, 156],
[161, 165, 199, 132],
[182, 192, 155, 178]])
>>> import numpy as np
>>> np.random.seed(10)
>>> np.random.randint(100,200,(3,4))
array([[109, 115, 164, 128],
[189, 193, 129, 108],
[173, 100, 140, 136]])
>>> np.random.randint(100,200,(3,4))
array([[116, 111, 154, 188],
[162, 133, 172, 178],
[149, 151, 154, 177]])
>>> np.random.seed(10)
>>> np.random.randint(100,200,(3,4))
array([[109, 115, 164, 128],
[189, 193, 129, 108],
[173, 100, 140, 136]])
| function | explain |
|---|---|
| shuffle(a) | Randomly arrange according to the first axis of array a and change array x |
| permutation(a) | Generate a new out of order array according to the first axis of array a without changing array x |
| choice(a[,size,replace,p]) | Extract elements from one-dimensional array a with probability p to form a new array of size shape. replace indicates whether elements can be reused. The default is True |
shuffle demo
>>> b=np.random.randint(100,200,(3,4))
>>> b
array([[164, 145, 135, 156],
[161, 165, 199, 132],
[182, 192, 155, 178]])
>>> a=np.random.randint(100,200,(3,4))
>>> a
array([[144, 104, 103, 126],
[156, 145, 190, 195],
[184, 186, 183, 160]])
>>> np.random.shuffle(a)
>>> a
array([[144, 104, 103, 126],
[184, 186, 183, 160],
[156, 145, 190, 195]])
>>> np.random.shuffle(a)
>>> a
array([[156, 145, 190, 195],
[184, 186, 183, 160],
[144, 104, 103, 126]])
permutation demo
>>> a=np.random.randint(100,200,(3,4))
>>> a
array([[138, 174, 192, 151],
[171, 109, 107, one hundred and twenty-seven],
[144, 174, 151, 138]])
>>> np.random.permutation(a)
array([[171, 109, 107, one hundred and twenty-seven],
[144, 174, 151, 138],
[138, 174, 192, 151]])
>>> a
array([[138, 174, 192, 151],
[171, 109, 107, 127],
[144, 174, 151, 138]])
choice demo
>>> b=np.random.randint(100,200,(8,))
>>> b
array([181, 106, 171, 103, 198, 168, 180, 192])
>>> np.random.choice(b,(3,2),replace=False)
array([[106, 168],
[198, 180],
[192, 103]])
>>> np.random.choice(b,(3,2),p=b/np.sum(b))
array([[168, 168],
[192, 171],
[192, 168]])
| function | explain |
|---|---|
| uniform(low,high,size) | Generate a uniformly distributed array, with low as the starting value, high as the ending value and size as the shape |
| normal(loc,scale,size) | Generate a normal distribution array, loc is the mean, scale is the standard deviation, and size is the shape |
| poisson(lam,size) | Generate a Poisson distribution array, lam is the probability of random events, and size is the shape |
>>> u=np.random.uniform(0,10,(3,4))
>>> u
array([[8.73861146, 0.46324114, 6.75838186, 0.71928162],
[5.3382006 , 5.31719917, 9.72423284, 0.74520933],
[2.51992423, 2.65729896, 1.41524432, 5.0901383 ]])
>>> n=np.random.normal(10,5,(3,4))
>>> n
array([[ 7.96208063, 20.35050238, 12.34165304, 6.72274216],
[24.59582282, 9.98507963, 9.87675547, 6.42073446],
[13.78549274, 7.68922186, 10.98222814, 5.73539134]])
Statistical function
| function | explain |
|---|---|
| sum(a,axis=None) | Calculate the sum of the related elements of array a according to the given axis, axis integer or tuple |
| mean(a,axis=None) | Calculate the related elements of array a according to the given axis, and expect axis integer or tuple |
| average(a,axis=None,weights=None) | Calculate the weighted average of the relevant elements of array a according to the given axis |
| std(a,axis=None) | Calculate the standard deviation of relevant elements of array a according to the given axis |
| var(a,axis=None) | Calculate the variance of related elements of array a according to the given axis |
>>> np.random.choice(b,(3,2),p=b/np.sum(b))
array([[168, 168],
[192, 171],
[192, 168]])
>>> u=np.random.uniform(0,10,(3,4))
>>> u
array([[8.73861146, 0.46324114, 6.75838186, 0.71928162],
[5.3382006 , 5.31719917, 9.72423284, 0.74520933],
[2.51992423, 2.65729896, 1.41524432, 5.0901383 ]])
>>> n=np.random.normal(10,5,(3,4))
>>> n
array([[ 7.96208063, 20.35050238, 12.34165304, 6.72274216],
[24.59582282, 9.98507963, 9.87675547, 6.42073446],
[13.78549274, 7.68922186, 10.98222814, 5.73539134]])
>>> a=np.arange(15).reshape(3,5)
>>> a
array([[ 0, 1, 2, 3, 4],
[ 5, 6, 7, 8, 9],
[10, 11, 12, 13, 14]])
>>> np.sum(a)
105
>>> np.mean(a,axis=1)
array([ 2., 7., 12.])
>>> np.mean(a,axis=0)
array([5., 6., 7., 8., 9.])
>>> np.average(a,axis=0,weights=[10,5,1])
array([2.1875, 3.1875, 4.1875, 5.1875, 6.1875])
>>> np.std(a)
4.320493798938574
>>> np.var(a)
18.666666666666668
| function | explain |
|---|---|
| min(a) max(a) | Calculates the minimum and maximum values of the elements in array a |
| argmin(a) argmax(a) | Calculate the subscript after one-dimensional reduction of the minimum and maximum values of elements in array a |
| unravel_index(index,shape) | Convert one-dimensional index into multi-dimensional index according to shape |
| ptp(a) | Calculates the difference between the maximum and minimum values of elements in array a |
| median(a) | Calculates the median of the elements in array a |
>>> b=np.arange(15,0,-1).reshape(3,5)
>>> b
array([[15, 14, 13, 12, 11],
[10, 9, 8, 7, 6],
[ 5, 4, 3, 2, 1]])
>>> np.max(b)
15
>>> np.argmax(b)
0
>>> np.unravel_index(np.argmax(b),b.shape)
(0, 0)
Gradient function
np.gradient(f)
Calculate the gradient of elements in array F. when f is multidimensional, return the gradient of each dimension
>>> a=np.random.randint(0,20,(5)) >>> a array([11, 3, 14, 18, 11]) >>> np.gradient(a) array([-8. , 1.5, 7.5, -1.5, -7. ])#1.5=(14-11)/2,-7=(11-18)/1 >>> b=np.random.randint(0,20,(5)) >>> b array([15, 12, 19, 7, 15]) >>> np.gradient(b) array([-3. , 2. , -2.5, -2. , 8. ])
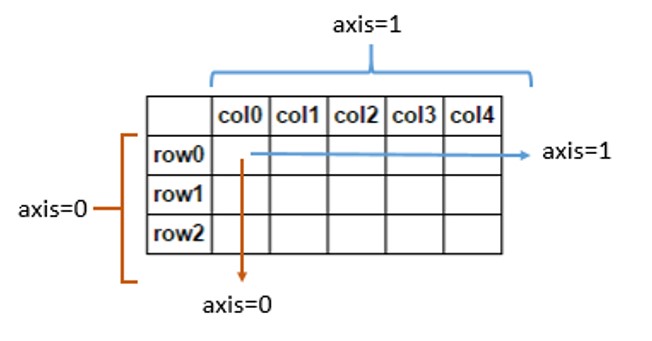
Understanding of axis
axis=0 removes the outermost parentheses and regards them as a whole. Operations are performed on this whole
axis=1 removes the second bracket and regards it as a whole. The operation is performed on this whole
... and so on
axis=0 means down, and axis=1 means across
- A value of 0 means that the method is executed down the label \ index value of each column or row
- A value of 1 indicates that the corresponding method is executed along the label direction of each row or column
Example: picture hand drawing effect
from PIL import Image
import numpy as np
a = np.asarray(Image.open('./beijing.jpg').convert('L')).astype('float')
depth = 10. # Preset depth value (0-100)
grad = np.gradient(a) #Take the gradient value of image gray level
grad_x, grad_y = grad #Take the gradient values of horizontal and vertical images respectively
grad_x = grad_x*depth/100.
grad_y = grad_y*depth/100. #Gradient normalization
A = np.sqrt(grad_x**2 + grad_y**2 + 1.)
uni_x = grad_x/A
uni_y = grad_y/A
uni_z = 1./A
vec_el = np.pi/2.2 # Top view angle of light source, radian value
vec_az = np.pi/4. # Azimuth angle of light source, radian value
dx = np.cos(vec_el)*np.cos(vec_az) #Influence of light source on x-axis
dy = np.cos(vec_el)*np.sin(vec_az) #Influence of light source on y-axis
dz = np.sin(vec_el) #Influence of light source on z-axis
b = 255*(dx*uni_x + dy*uni_y + dz*uni_z) #Light source normalization
b = b.clip(0,255)
im = Image.fromarray(b.astype('uint8')) #Reconstructed image
im.save('./beijingHD.jpg')
2. Matlab Library
Basic operation
import matplotlib.pyplot as plt
plt.plot([0,2,4,6,8],[3,1,4,5,2])#If there is only one, it is regarded as the ordinate
plt.ylabel("grade")#Label
plt.savefig('test',dpi=600)#Store pictures
plt.axis([-1,10,0,6])#Horizontal axis range
plt.subplot(3,2,4)#It is divided into 3 * 2 sub drawing space and painted in the fourth space
plt.show()
plot() function
plt.plot(x,y,format_string,**kwargs)
- x: X-axis data, list or array, can not be written
- y: Y-axis data, list or array, must have
- format_string: format string of control curve; optional
- **kwargs: second group or more (x, y, format)_ string)
- When drawing multiple curves, the x of each curve cannot be omitted
>>> import matplotlib.pyplot as plt >>> import numpy as np >>> a=np.arange(10) >>> plt.plot(a,a*1.5,a,a*2.5,a,a*3.5,a,a*4.5) [<matplotlib.lines.Line2D object at 0x000002BABE2752E0>, <matplotlib.lines.Line2D object at 0x000002BABE275160>, <matplotlib.lines.Line2D object at 0x000002BABE275460>, <matplotlib.lines.Line2D object at 0x000002BABE2754F0>] >>> plt.show()

format_string value: color character + style character + mark character
| Color character | explain | Color character | explain |
|---|---|---|---|
| 'b' | blue | 'm' | Magenta |
| 'g' | green | 'y' | yellow |
| 'r' | gules | 'k' | black |
| 'c' | Turquoise | 'w' | white |
| '#008000' | RGB a color | '0.8' | Grayscale value string |
| Style character | explain |
|---|---|
| '-' | Solid line |
| '–' | Broken broken line |
| '-.' | Dotted line |
| ':' | Dotted line |
| '''' | Wireless strip |
| Marker character | explain | Marker character | explain | Marker character | explain |
|---|---|---|---|---|---|
| '.' | Point marker | '1' | Lower flower triangle mark | 'h' | Vertical hexagon mark |
| ',' | Pixel marker | '2' | Flower triangle mark | 'H' | Horizontal hexagon mark |
| 'o' | Solid circle mark | '3' | Left flower triangle mark | '+' | Cross mark |
| 'v' | Inverted triangle mark | '4' | Right flower triangle mark | 'x' | x mark |
| '^' | Upper triangle mark | 's' | Filled square marker | 'D' | Diamond mark |
| '>' | Right triangle mark | 'p' | Black pentagonal marker | 'd' | Thin diamond marker |
| '<' | Left triangle mark | '*' | Star Mark | '|' | Vertical line marking |
>>> import matplotlib.pyplot as plt >>> import numpy as np >>> a=np.arange(10) >>> plt.plot(a,a*1.5,'go-',a,a*2.5,'rx',a,a*3.5,'*',a,a*4.5,'b-.') [<matplotlib.lines.Line2D object at 0x000002BABAC37670>, <matplotlib.lines.Line2D object at 0x000002BABAC37730>, <matplotlib.lines.Line2D object at 0x000002BABAC37610>, <matplotlib.lines.Line2D object at 0x000002BABAC377C0>] >>> plt.show()

**kwargs
- Color: control color, color = "green"
- linstyle: line style, linestyle = "dashed"
- Marker: marker style, marker = 'o'
- Markerfacecolor: marker color, markerfacecolor = 'blue'
- Markersize: mark size, markersize=20
Chinese display (recommendation 2)
pylot does not support Chinese display by default
- Rcparams (Global modification)
| attribute | explain |
|---|---|
| 'font.family' | The name used to display the font |
| 'font.style' | The font style is normal, italic; italic |
| 'font.size' | Font size, integer font size or 'large', 'x-small' |
| Chinese font | explain |
|---|---|
| 'SimHei' | Chinese bold |
| 'Kaiti' | Chinese regular script |
| 'LiSu' | Chinese official script |
| 'FangSong' | Chinese imitation Song Dynasty |
| 'YouYuan' | Chinese immature circle |
| 'STSong' | Chinese Song typeface |
import matplotlib.pyplot as plt
import matplotlib
matplotlib.rcParams['font.family']='SimHei'
plt.plot([3,1,4,5,2])
plt.ylabel("Longitudinal axis (value)")
plt.savefig('test',dpi=600)
plt.show()

import matplotlib.pyplot as plt
import matplotlib
import numpy as np
matplotlib.rcParams['font.family']='Microsoft YaHei'
matplotlib.rcParams['font.size']=20
a=np.arange(0.0,5.0,0.02)
plt.xlabel('Horizontal axis: time')
plt.ylabel('Longitudinal axis: amplitude')
plt.plot(a,np.cos(2*np.pi*a),'r--')
plt.show()

- Fontproperties (local modification)
import matplotlib.pyplot as plt
import numpy as np
a=np.arange(0.0,5.0,0.02)
plt.xlabel('Horizontal axis: time',fontproperties='SimHei',fontsize=20)
plt.ylabel('Longitudinal axis: amplitude',fontproperties='SimHei',fontsize=20)
plt.plot(a,np.cos(2*np.pi*a),'r--')
plt.show()

Text display
| function | explain |
|---|---|
| plt.xlabel() | Add a text label to the x-axis |
| plt.ylabel() | Add a text label to the y-axis |
| plt.title() | Add text labels to the graphics as a whole |
| plt.text() | Add text anywhere |
| plt.annotate() | Add notes with arrows in the drawing |
import matplotlib.pyplot as plt
import numpy as np
a=np.arange(0.0,5.0,0.02)
plt.plot(a,np.cos(2*np.pi*a),'r--')
plt.xlabel('Horizontal axis: time',fontproperties='SimHei',fontsize=20,color='green')
plt.ylabel('Longitudinal axis: amplitude',fontproperties='SimHei',fontsize=20)
plt.title(r'Sine wave example $y=cos(2\pi x)$',fontproperties='SimHei',fontsize=20)
plt.text(2,1,r'$\mu=100$',fontsize=15)
plt.axis([-1,6,-2,2])
plt.grid(True)
plt.show()

import matplotlib.pyplot as plt
import numpy as np
a=np.arange(0.0,5.0,0.02)
plt.plot(a,np.cos(2*np.pi*a),'r--')
plt.xlabel('Horizontal axis: time',fontproperties='SimHei',fontsize=25,color='green')
plt.ylabel('Longitudinal axis: amplitude',fontproperties='SimHei',fontsize=25)
plt.title(r'Sine wave example $y=cos(2\pi x)$',fontproperties='SimHei',fontsize=25)
plt.annotate(r'$\mu=100$',xy=(2,1),xytext=(3,1.5),arrowprops=dict(facecolor='black',shrink=0.1,width=2))
plt.axis([-1,6,-2,2])
plt.grid(True)
plt.show()

Sub drawing area
plt.subplot2grid(GridSpec,CurSpec,colspan=1,rowspan=1)
-
GridSpec is a tuple that describes the partition of the sketchpad
-
CurSpec is a tuple, making a grid
-
colspan represents the extension of the column
-
rowspan represents an extension of a row
plt.subplot2grid((3,3),(0,0),colspan=3) plt.subplot2grid((3,3),(1,0),colspan=2) plt.subplot2grid((3,3),(1,2),rowspan=2) plt.subplot2grid((3,3),(2,0)) plt.subplot2grid((3,3),(2,1))

import matplotlib.gridspec as gridspec import matplotlib.pyplot as plt gs=gridspec.GridSpec(3,3) ax1=plt.subplot(gs[0:,]) ax2=plt.subplot(gs[1:-1]) ax3=plt.subplot(gs[1:,-1]) ax4=plt.subplot(gs[2,0]) ax5=plt.subplot(gs[2,1])
Basic chart function
| function | explain |
|---|---|
| plt.plot(x,y,fmt,...) | Coordinate diagram |
| plt.boxplot(data,notch,position) | Box diagram |
| plt.bar(left,height,width,bottom) | Bar chart |
| plt.barh(width,bottom,left,height) | Horizontal bar chart |
| plt.polar(theta,r) | Polar diagram |
| plt.pie(data,explode) | Pie chart |
| plt.psd(x,NFFT=256,pad_to,F) | Power spectral density diagram |
| plt.specgram(x,NFFT=256,pad_to.F) | Spectrogram |
| plt.cohere(x,y,NFFT=256,Fs) | x-y correlation function |
| plt.scatter(x,y) | Scatter plot where xy is the same length |
| plt.step(x,y,where) | Step diagram |
| plt.hist(x,bins,normed) | histogram |
- Pie chart
import matplotlib.pyplot as pltlabels='frogs','hogs','dogs','logs'size=[15,30,45,10]explode=(0,0.1,0,0)plt.pie(size,explode=explode,labels=labels,autopct='%1.1f%%',shadow=False,startangle=90)plt.axis('equal')plt.show()

- histogram
import matplotlib.pyplot as pltimport numpy as npnp.random.seed(0)mu,sigma=100,20a=np.random.normal(mu,sigma,size=100)plt.hist(a,20,density=True,histtype='stepfilled',facecolor='b',alpha=0.75)#density normalization plt.title('Histogram')plt.show()

- Polar diagram
import matplotlib.pyplot as pltimport numpy as npN=20theta=np.linspace(0.0,2*np.pi,N,endpoint=False)radii=10*np.random.rand(N)width=np.pi/4*np.random.rand(N)ax=plt.subplot(111,projection='polar')#Create polar subgraph bars=ax.bar(theta,radii,width=width,bottom=0.0)#theta, radio, width corresponds to left, height, width for R, bar in zip (radio, bars): bar.set_ facecolor(plt.cm.viridis(r/10.)) bar.set_ alpha(0.5)plt.show()

- Scatter diagram
import matplotlib.pyplot as pltimport numpy as npfig,ax=plt.subplots()ax.plot(10*np.random.randn(100),10*np.random.randn(100),'o')ax.set_title('Simple Scatter')plt.show()

3.Pandas Library
Two data types: series and dataframe
Series type
Custom index
It is composed of a set of data and its related indexes. It allows user-defined indexes
>>> import pandas as pd >>> a=pd.Series([9,8,7,6]) >>> a 0 9 1 8 2 7 3 6 dtype: int64 >>> b=pd.Series([9,8,7,6],index=['a','b','c','d']) >>> b a 9 b 8 c 7 d 6 dtype: int64
establish
-
Python list
-
Scalar value
>>> s=pd.Series(25,index=['a','b','c'])#The index here cannot be omitted >>> s a 25 b 25 c 25 dtype: int64
-
Python dictionary
>>> d=pd.Series({'a':9,'b':8,'c':7}) >>> d a 9 b 8 c 7 dtype: int64 >>> e=pd.Series({'a':9,'b':8,'c':7},index=['c','a','b','d'])#Select from the dictionary >>> e c 7.0 a 9.0 b 8.0 d NaN dtype: float64 -
ndarray
>>> import pandas as pd >>> import numpy as np >>> n=pd.Series(np.arange(5)) >>> n 0 0 1 1 2 2 3 3 4 4 dtype: int32 >>> m=pd.Series(np.arange(5),index=np.arange(9,4,-1)) >>> m 9 0 8 1 7 2 6 3 5 4 dtype: int32
-
Other functions
basic operation
Similar to ndarray, dictionary
>>> import pandas as pd >>> b=pd.Series([9,8,7,6],['a','b','c','d']) >>> b a 9 b 8 c 7 d 6 dtype: int64 >>> b.index Index(['a', 'b', 'c', 'd'], dtype='object') >>> b.values array([9, 8, 7, 6], dtype=int64)
>>> b['b']8>>> b[1]8>>> b[['c','d',0]]#Either of the two lassos can be referenced, but cannot be mixed > > > b [['C','d ',' a ']]c 7d 6a 9dtype: Int64
The Series operation is similar to the ndarray type
- The index method is the same, []
- Operations and operations in Numpy are available for Series types
- You can slice through a list of custom indexes
- It can be sliced through automatic index. If there is a custom index, it will be sliced together
>>> import pandas as pd>>> b=pd.Series([9,8,7,6],['a','b','c','d'])>>> ba 9b 8c 7d 6dtype: int64>>> b[3]6>>> b[:3]a 9b 8c 7dtype: int64>>> b[b>b.median()]a 9b 8dtype: int64>>> np.exp(b)a 8103.083928b 2980.957987c 1096.633158d 403.428793dtype: float64
The Series type operation is similar to the Python dictionary type
- Access through custom index
- Reserved word in operation
- Use the. get() method
>>> import pandas as pd>>> b=pd.Series([9,8,7,6],['a','b','c','d'])>>> b['b']8>>> 'c' in bTrue>>> 0 in bFalse>>> b.get('f',100)100
Series type alignment operation
>>> import pandas as pd>>> a=pd.Series([1,2,3],['c','d','e'])>>> b=pd.Series([9,8,7,6],['a','b','c','d'])>>> a+ba NaNb NaNc 8.0d 8.0e NaNdtype: float64#The Series type automatically aligns the data of different indexes during operation
name attribute of Series type
>>> import pandas as pd>>> b=pd.Series([9,8,7,6],['a','b','c','d'])>>> b.name>>> b.name='Series object'>>> b.index.name='Index column'>>> b Index column a 9b 8c 7d 6Name: Series object, dtype: int64
Series objects can be modified at any time and take effect immediately
>>> import pandas as pd>>> b=pd.Series([9,8,7,6],['a','b','c','d'])>>> b['a']=15>>> b.name="series">>> ba 15b 8c 7d 6Name: series, dtype: int64>>> b.name="New Series">>> b['b','c']=20>>> ba 15b 20c 20d 6Name: New Series, dtype: int64
DataFrame type
Consists of a set of columns that share an index
Row index, column index, column index
Cross column operation axis=1
Cross row operation axis=0
establish
- 2D ndarray object
>>> import pandas as pd>>> import numpy as np>>> d=pd.DataFrame(np.arange(10).reshape(2,5))>>> d 0 1 2 3 4#Automatic column index, 01 on the left is automatic row index 0 0 0 1 2 3 41 5 6 7 8 9
- A dictionary consisting of one-dimensional ndarray, list, dictionary, tuple, or Series
>>> import pandas as pd>>> dt={'one':pd.Series([1,2,3],index=['a','b','c']),'two':pd.Series([9,8,7,6],index=['a','b','c','d'])}>>> d=pd.DataFrame(dt)>>> d one two#Custom column index and custom row index a 1.0 9b 2.0 8c 3.0 7d NaN 6>>> pd.DataFrame(dt,index=['b','c','d'],columns=['two','three']) two three#The data is automatically supplemented according to the row index b 8 NANC 7 NAND 6 Nan
- Create from a list type dictionary
>>> import pandas as pd>>> dl={'one':[1,2,3,4],'two':[9,8,7,6]}>>> d=pd.DataFrame(dl,index=['a','b','c','d'])>>> d one twoa 1 9b 2 8c 3 7d 4 6
example
>>> import pandas as pd>>> dl={'city':['Beijing','Shanghai','Guangzhou','Shenzhen','Shenyang'], 'Ring ratio':[101.5,101.2,101.3,102.0,100.1], 'Year on year':[120.7,127.3,119.4,140.9,101.4], 'Fixed base':[121.4,127.8,120.0,145.5,101.6]}>>> d=pd.DataFrame(dl,index=['c1','c2','c3','c4','c5'])>>> d city Ring ratio Year on year Fixed base c1 Beijing 101.5 120.7 121.4c2 Shanghai 101.2 127.3 127.8c3 Guangzhou 101.3 119.4 120.0c4 Shenzhen 102.0 140.9 145.5c5 Shenyang 100.1 101.4 101.6>>> d.indexIndex(['c1', 'c2', 'c3', 'c4', 'c5'], dtype='object')>>> d.columnsIndex(['city', 'Ring ratio', 'Year on year', 'Fixed base'], dtype='object')>>> d.valuesarray([['Beijing', 101.5, 120.7, 121.4], ['Shanghai', 101.2, 127.3, 127.8], ['Guangzhou', 101.3, 119.4, 120.0], ['Shenzhen', 102.0, 140.9, 145.5], ['Shenyang', 100.1, 101.4, 101.6]], dtype=object)>>> d['Year on year']c1 120.7c2 127.3c3 119.4c4 140.9c5 101.4Name: Year on year, dtype: float64>>> d.loc['c2']city Shanghai Chain comparison 101.2 Year on year 127.3 Fixed base 127.8Name: c2, dtype: object>>> d['Year on year']['c2']127.3
- Series
- Other dataframes
operation
reindex
.reindex()Capable of changing or remaking Series and DataFrame Index of
>>> import pandas as pd>>> dl={'city':['Beijing','Shanghai','Guangzhou','Shenzhen','Shenyang'],'Ring ratio':[101.5,101.2,101.3,102.0,100.1],'Year on year':[120.7,127.3,119.4,140.9,101.4],'Fixed base':[121.4,127.8,120.0,145.5,101.6]}>>> d=pd.DataFrame(dl,index=['c1','c2','c3','c4','c5'])>>> d city Ring ratio Year on year Fixed base c1 Beijing 101.5 120.7 121.4c2 Shanghai 101.2 127.3 127.8c3 Guangzhou 101.3 119.4 120.0c4 Shenzhen 102.0 140.9 145.5c5 Shenyang 100.1 101.4 101.6>>> d=d.reindex(index=['c5','c4','c3','c2','c1'])>>> d city Ring ratio Year on year Fixed base c5 Shenyang 100.1 101.4 101.6c4 Shenzhen 102.0 140.9 145.5c3 Guangzhou 101.3 119.4 120.0c2 Shanghai 101.2 127.3 127.8c1 Beijing 101.5 120.7 121.4>>> d=d.reindex(columns=['city','Year on year','Ring ratio','Fixed base'])>>> d city Year on year Ring ratio Fixed base c5 Shenyang 101.4 100.1 101.6c4 Shenzhen 140.9 102.0 145.5c3 Guangzhou 119.4 101.3 120.0c2 Shanghai 127.3 101.2 127.8c1 Beijing 120.7 101.5 121.4
.reindex(index=None,columns=None,...)
| parameter | explain |
|---|---|
| index,columns | New row column custom index |
| fill_value | In the re index, the value used to populate the missing location |
| method | Filling method, fill forward with fill and fill backward with bfill |
| limit | Maximum fill |
| copy | The default is True to generate a new object; When False, the old and new equality is not copied |
>>> newc=d.columns.insert(4,'newly added')>>> newd=d.reindex(columns=newc,fill_value=200)>>> newd city Year on year Ring ratio Fixed base addition c5 Shenyang 101.4 100.1 101.6 200c4 Shenzhen 140.9 102.0 145.5 200c3 Guangzhou 119.4 101.3 120.0 200c2 Shanghai 127.3 101.2 127.8 200c1 Beijing 120.7 101.5 121.4 200
Index type operation
Index type: the indexes of Series and DataFrame are of index type, and the index object is of non modifiable type
Common methods of index type
| method | explain |
|---|---|
| .append(idx) | Connect another Index object to generate a new Index object |
| .diff(idx) | Calculate the difference set to generate a new Index object |
| .intersection(idx) | Calculate intersection |
| union(idx) | Computational Union |
| .delete(loc) | Delete the element at loc location |
| .insert(loc,e) | Add an element e at loc |
>>> d
city Year on year Ring ratio Fixed base
c5 Shenyang 101.4 100.1 101.6
c4 Shenzhen 140.9 102.0 145.5
c3 Guangzhou 119.4 101.3 120.0
c2 Shanghai 127.3 101.2 127.8
c1 Beijing 120.7 101.5 121.4
>>> nc=d.columns.delete(2)
>>> ni=d.index.insert(5,'c0')
>>> nd=d.reindex(index=ni,columns=nc,method='ffill')#Will report an error
>>> nd=d.reindex(index=ni,columns=nc).ffill()
>>> nd
city Year on year Fixed base
c5 Shenyang 101.4 101.6
c4 Shenzhen 140.9 145.5
c3 Guangzhou 119.4 120.0
c2 Shanghai 127.3 127.8
c1 Beijing 120.7 121.4
c0 Beijing 120.7 121.4
Deletes the specified index object
. drop() can delete the row or column indexes specified by Series and DataFrame
>>> a=pd.Series([9,8,7,6],index=['a','b','c','d'])
>>> a
a 9
b 8
c 7
d 6
dtype: int64
>>> a.drop(['b','c'])
a 9
d 6
dtype: int64
>>> d
city Year on year Ring ratio Fixed base
c5 Shenyang 101.4 100.1 101.6
c4 Shenzhen 140.9 102.0 145.5
c3 Guangzhou 119.4 101.3 120.0
c2 Shanghai 127.3 101.2 127.8
c1 Beijing 120.7 101.5 121.4
>>> d.drop('c5')
city Year on year Ring ratio Fixed base
c4 Shenzhen 140.9 102.0 145.5
c3 Guangzhou 119.4 101.3 120.0
c2 Shanghai 127.3 101.2 127.8
c1 Beijing 120.7 101.5 121.4
>>> d.drop('Year on year',axis=1)
city Ring ratio Fixed base
c5 Shenyang 100.1 101.6
c4 Shenzhen 102.0 145.5
c3 Guangzhou 101.3 120.0
c2 Shanghai 101.2 127.8
c1 Beijing 101.5 121.4
Data type operation
- add , subtract , multiply and divide
>>> a=pd.DataFrame(np.arange(12).reshape(3,4))
>>> a
0 1 2 3
0 0 1 2 3
1 4 5 6 7
2 8 9 10 11
>>> b=pd.DataFrame(np.arange(20).reshape(4,5))
>>> b
0 1 2 3 4
0 0 1 2 3 4
1 5 6 7 8 9
2 10 11 12 13 14
3 15 16 17 18 19
>>> a+b
0 1 2 3 4
0 0.0 2.0 4.0 6.0 NaN
1 9.0 11.0 13.0 15.0 NaN
2 18.0 20.0 22.0 24.0 NaN
3 NaN NaN NaN NaN NaN
>>> a*b
0 1 2 3 4
0 0.0 1.0 4.0 9.0 NaN
1 20.0 30.0 42.0 56.0 NaN
2 80.0 99.0 120.0 143.0 NaN
3 NaN NaN NaN NaN NaN
Operation in method form
| method | explain |
|---|---|
| .add(d,**argws) | Addition between types, optional parameters |
| .sub(d,**argws) | Inter type subtraction, optional parameters |
| .mul(d,**argws) | Inter type multiplication, optional parameters |
| .div(d,**argws) | Division between types, optional parameters |
>>> a
0 1 2 3
0 0 1 2 3
1 4 5 6 7
2 8 9 10 11
>>> b
0 1 2 3 4
0 0 1 2 3 4
1 5 6 7 8 9
2 10 11 12 13 14
3 15 16 17 18 19
>>> b.add(a,fill_value=100)#Replace NAN first and then participate in the operation
0 1 2 3 4
0 0.0 2.0 4.0 6.0 104.0
1 9.0 11.0 13.0 15.0 109.0
2 18.0 20.0 22.0 24.0 114.0
3 115.0 116.0 117.0 118.0 119.0
>>> a.mul(b,fill_value=0)
0 1 2 3 4
0 0.0 1.0 4.0 9.0 0.0
1 20.0 30.0 42.0 56.0 0.0
2 80.0 99.0 120.0 143.0 0.0
3 0.0 0.0 0.0 0.0 0.0
Broadcast operation is performed between different dimensions, and one-dimensional Series participates in the operation on axis 1 by default
>>> b
0 1 2 3 4
0 0 1 2 3 4
1 5 6 7 8 9
2 10 11 12 13 14
3 15 16 17 18 19
>>> c=pd.Series(np.arange(4))
>>> c
0 0
1 1
2 2
3 3
dtype: int32
>>> c-10
0 -10
1 -9
2 -8
3 -7
dtype: int32
>>> b-c#axis=1, cross column
0 1 2 3 4
0 0.0 0.0 0.0 0.0 NaN
1 5.0 5.0 5.0 5.0 NaN
2 10.0 10.0 10.0 10.0 NaN
3 15.0 15.0 15.0 15.0 NaN
Make one-dimensional Series participate in axis 0 operation
>>> b
0 1 2 3 4
0 0 1 2 3 4
1 5 6 7 8 9
2 10 11 12 13 14
3 15 16 17 18 19
>>> c
0 0
1 1
2 2
3 3
dtype: int32
>>> b.sub(c,axis=0)
0 1 2 3 4
0 0 1 2 3 4
1 4 5 6 7 8
2 8 9 10 11 12
3 12 13 14 15 16
- Comparison operation
>>> a=pd.DataFrame(np.arange(12).reshape(3,4))
>>> d=pd.DataFrame(np.arange(12,0,-1).reshape(3,4))
>>> a
0 1 2 3
0 0 1 2 3
1 4 5 6 7
2 8 9 10 11
>>> d
0 1 2 3
0 12 11 10 9
1 8 7 6 5
2 4 3 2 1
>>> a>d
0 1 2 3
0 False False False False
1 False False False True
2 True True True True
>>> a==d
0 1 2 3
0 False False False False
1 False False True False
2 False False False False
>>> a
0 1 2 3
0 0 1 2 3
1 4 5 6 7
2 8 9 10 11
>>> c
0 0
1 1
2 2
3 3
dtype: int32
>>> a>c
0 1 2 3
0 False False False False
1 True True True True
2 True True True True
>>> c>0
0 False
1 True
2 True
3 True
dtype: bool
Characteristic analysis of pandas data
.sort_ The index () method sorts by index on the specified axis. The default is ascending
.sort_index(axis=0,ascending=True)
>>> b=pd.DataFrame(np.arange(20).reshape(4,5),index=['c','a','d','b'])
>>> b
0 1 2 3 4
c 0 1 2 3 4
a 5 6 7 8 9
d 10 11 12 13 14
b 15 16 17 18 19
>>> b.sort_index()
0 1 2 3 4
a 5 6 7 8 9
b 15 16 17 18 19
c 0 1 2 3 4
d 10 11 12 13 14
>>> b.sort_index(ascending=False)
0 1 2 3 4
d 10 11 12 13 14
c 0 1 2 3 4
b 15 16 17 18 19
a 5 6 7 8 9
>>> b
0 1 2 3 4
c 0 1 2 3 4
a 5 6 7 8 9
d 10 11 12 13 14
b 15 16 17 18 19
>>> c=b.sort_index(axis=1,ascending=False)
>>> c
4 3 2 1 0
c 4 3 2 1 0
a 9 8 7 6 5
d 14 13 12 11 10
b 19 18 17 16 15
>>> c=c.sort_index()
>>> c
4 3 2 1 0
a 9 8 7 6 5
b 19 18 17 16 15
c 4 3 2 1 0
d 14 13 12 11 10
.sort_ The values () method sorts values on the specified axis. The default is ascending
Series.sort_values(axis=0,ascending=True)DataFrame.sort_values(by,axis=0,ascending=True)
- by: an index or index list on the axis
>>> b 0 1 2 3 4c 0 1 2 3 4a 5 6 7 8 9d 10 11 12 13 14b 15 16 17 18 19>>> c=b.sort_values(2,ascending=False)>>> c 0 1 2 3 4b 15 16 17 18 19d 10 11 12 13 14a 5 6 7 8 9c 0 1 2 3 4>>> c=c.sort_values('a',axis=1,ascending=False)>>> c 4 3 2 1 0b 19 18 17 16 15d 14 13 12 11 10a 9 8 7 6 5c 4 3 2 1 0
NaN put it last
Basic statistical analysis
For Series and DataFrame
| method | explain |
|---|---|
| .sum() | Calculate the sum of data according to the 0 axis |
| .count() | Number of non NaN values |
| .mean() .median() | Calculate the arithmetic mean and arithmetic median of the data |
| .var() .std() | Calculate the variance and standard deviation of the data |
| .min() .max() | Calculate the minimum and maximum values of the data |
Series only
| method | explain |
|---|---|
| .argmin() .argmax() | Calculate the automatic index of the location where the maximum and minimum values of data are located |
| .idxmin() .idxmax() | Calculate the user-defined index where the maximum and minimum values of data are located |
For Series and DataFrame types
| method | explain |
|---|---|
| .describe() | Statistical summary for axis 0 (columns) |
>>> a=pd.Series([9,8,7,6],index=['a','b','c','d']) >>> a a 9 b 8 c 7 d 6 dtype: int64 >>> a.describe() count 4.000000 mean 7.500000 std 1.290994 min 6.000000 25% 6.750000 50% 7.500000 75% 8.250000 max 9.000000 dtype: float64 >>> type(a.describe()) <class 'pandas.core.series.Series'> >>> a.describe()['count'] 4.0 >>> a.describe()['max'] 9.0
>>> b
0 1 2 3 4
c 0 1 2 3 4
a 5 6 7 8 9
d 10 11 12 13 14
b 15 16 17 18 19
>>> b.describe()
0 1 2 3 4
count 4.000000 4.000000 4.000000 4.000000 4.000000
mean 7.500000 8.500000 9.500000 10.500000 11.500000
std 6.454972 6.454972 6.454972 6.454972 6.454972
min 0.000000 1.000000 2.000000 3.000000 4.000000
25% 3.750000 4.750000 5.750000 6.750000 7.750000
50% 7.500000 8.500000 9.500000 10.500000 11.500000
75% 11.250000 12.250000 13.250000 14.250000 15.250000
max 15.000000 16.000000 17.000000 18.000000 19.000000
>>> type(b.describe())
<class 'pandas.core.frame.DataFrame'>
>>> b.describe().loc['max']
0 15.0
1 16.0
2 17.0
3 18.0
4 19.0
Name: max, dtype: float64
>>> b.describe()[2]
count 4.000000
mean 9.500000
std 6.454972
min 2.000000
25% 5.750000
50% 9.500000
75% 13.250000
max 17.000000
Name: 2, dtype: float64
Cumulative statistical analysis
| method | explain |
|---|---|
| .cumsum() | The sum of the first 1, 2... n numbers is given in turn |
| .cumprod() | The product of the first 1, 2... n numbers is given in turn |
| .cummax() | The maximum values of the first 1, 2... n numbers are given in turn |
| .cummin() | The minimum values of the first 1, 2... n numbers are given in turn |
>>> b
0 1 2 3 4
c 0 1 2 3 4
a 5 6 7 8 9
d 10 11 12 13 14
b 15 16 17 18 19
>>> b.cumsum()
0 1 2 3 4
c 0 1 2 3 4
a 5 7 9 11 13
d 15 18 21 24 27
b 30 34 38 42 46
>>> b.cumprod()
0 1 2 3 4
c 0 1 2 3 4
a 0 6 14 24 36
d 0 66 168 312 504
b 0 1056 2856 5616 9576
>>> b.cummin()
0 1 2 3 4
c 0 1 2 3 4
a 0 1 2 3 4
d 0 1 2 3 4
b 0 1 2 3 4
>>> b.cummax()
0 1 2 3 4
c 0 1 2 3 4
a 5 6 7 8 9
d 10 11 12 13 14
b 15 16 17 18 19
Applicable to Series and DataFrame types, rolling calculation (window calculation)
| method | explain |
|---|---|
| .rolling(w).sum() | Calculate the sum of adjacent w elements in turn |
| .rolling(w).mean() | Calculate the arithmetic mean of adjacent w elements in turn |
| .rolling(w).var() | Calculate the variance of adjacent w elements in turn |
| .rolling(w).std() | Calculate the standard deviation of adjacent w elements in turn |
| .rolling(w).min() .max() | Calculate the minimum and maximum values of adjacent w elements in turn |
>>> b
0 1 2 3 4
c 0 1 2 3 4
a 5 6 7 8 9
d 10 11 12 13 14
b 15 16 17 18 19
>>> b.rolling(2).sum()
0 1 2 3 4
c NaN NaN NaN NaN NaN
a 5.0 7.0 9.0 11.0 13.0
d 15.0 17.0 19.0 21.0 23.0#21=8+13
b 25.0 27.0 29.0 31.0 33.0
>>> b.rolling(3).sum()
0 1 2 3 4
c NaN NaN NaN NaN NaN
a NaN NaN NaN NaN NaN
d 15.0 18.0 21.0 24.0 27.0
b 30.0 33.0 36.0 39.0 42.0#42=9+14+19
correlation analysis
Pearson correlation coefficient
r
=
∑
i
=
1
n
(
x
i
−
x
‾
)
(
y
i
−
y
‾
)
∑
i
=
1
n
(
x
i
−
x
‾
)
2
∑
i
=
1
n
(
y
i
−
y
‾
)
2
r=\frac{\sum_{i=1}^n(x_i-\overline x)(y_i-\overline y)}{\sqrt{\sum _{i=1}^n(x_i-\overline x)^2}\sqrt{\sum _{i=1}^n(y_i-\overline y)^2}}
r=∑i=1n(xi−x)2
∑i=1n(yi−y)2
∑i=1n(xi−x)(yi−y)
| correlation coefficient | Degree of correlation |
|---|---|
| 0.8-1.0 | Extremely strong correlation |
| 0.6-0.8 | Strong correlation |
| 0.4-0.6 | Moderate correlation |
| 0.2-0.4 | Weak correlation |
| 0.0-0.2 | Very weak correlation or no correlation |
For Series and DataFrame types
| method | explain |
|---|---|
| .cov() | Calculate covariance matrix |
| .corr() | Calculate correlation coefficient matrix |
>>> import pandas as pd >>> hprice=pd.Series([3.04,22.93,12.75,22.6,12.33],index=['2008','2009','2010','2011','2012']) >>> m2=pd.Series([8.18,18.38,9.13,7.82,6.69],index=['2008','2009','2010','2011','2012']) >>> hprice.corr(m2) 0.5239439145220387