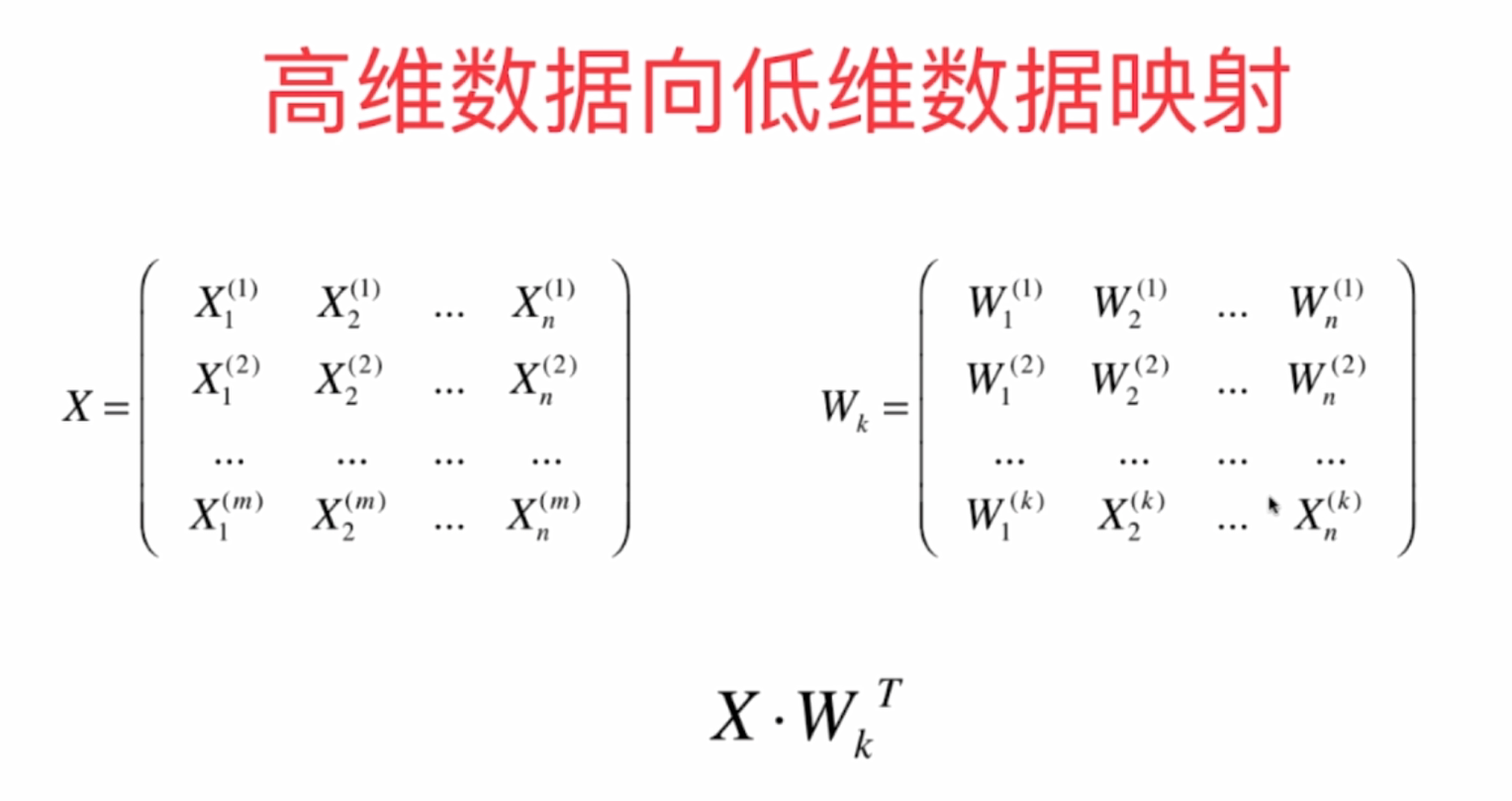
1 Basic Concepts
- PCA is principal component analysis. Principal component analysis, also known as principal component analysis, aims to use the idea of dimensionality reduction to transform multiple indicators into a few comprehensive indicators.
- In statistics, PCA is a technique to simplify data sets. It's a linear transformation. This transformation transforms the data into a new coordinate system, so that the first variance of any data projection is on the first coordinate (called the first principal component), the second variance is on the second coordinate (the second principal component), and so on.
- Principal component analysis (PCA) is often used to reduce the dimension of data sets, while maintaining the feature of the largest contribution of the opposite difference. This is achieved by retaining the low-order principal components and ignoring the high-order principal components. In this way, low-order components can often retain the most important aspects of the data. However, this is not certain. It depends on the specific application.
2 principle and mathematical derivation
1. The gradient rise method is used in principal component analysis.
Characteristic
Principle:
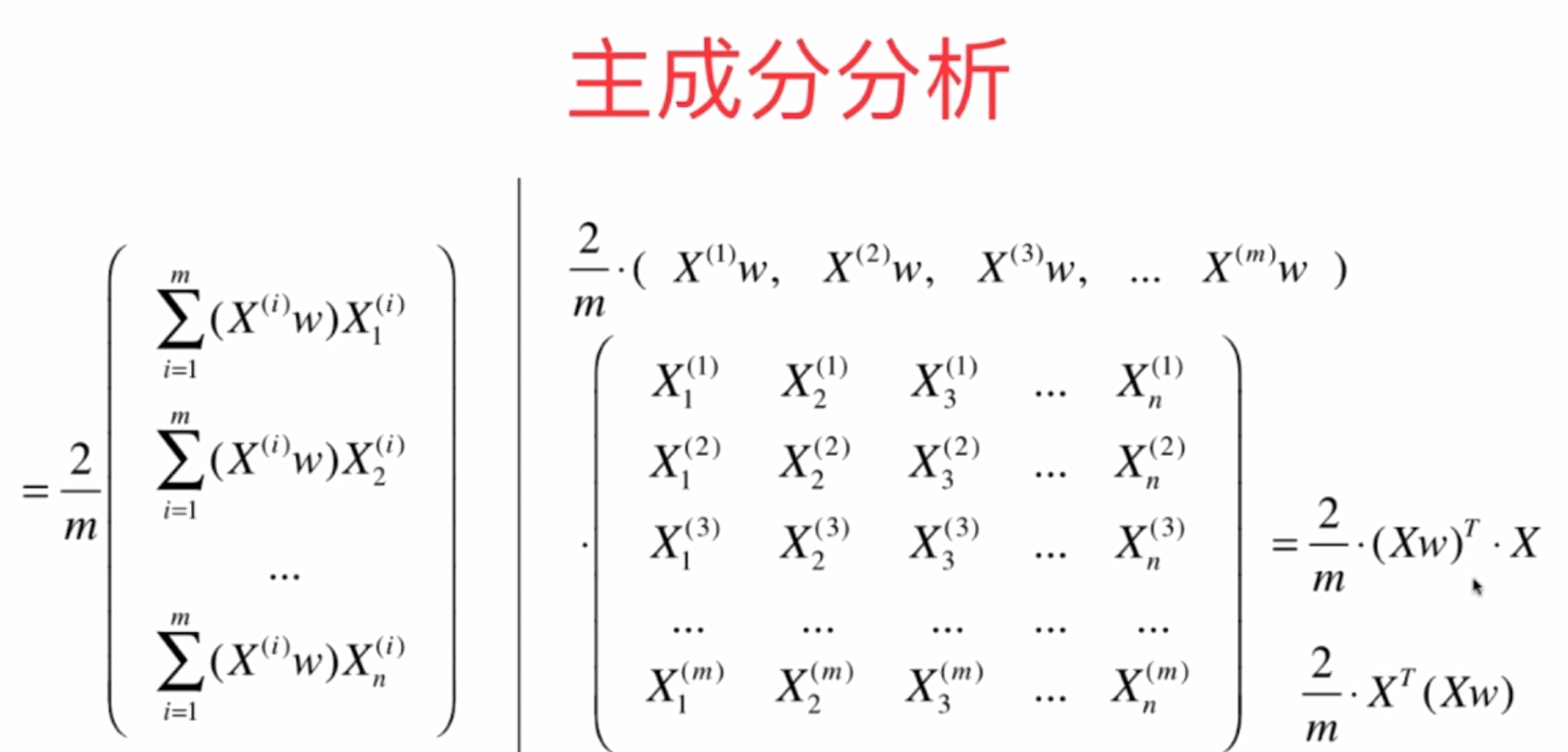
Mathematical derivation: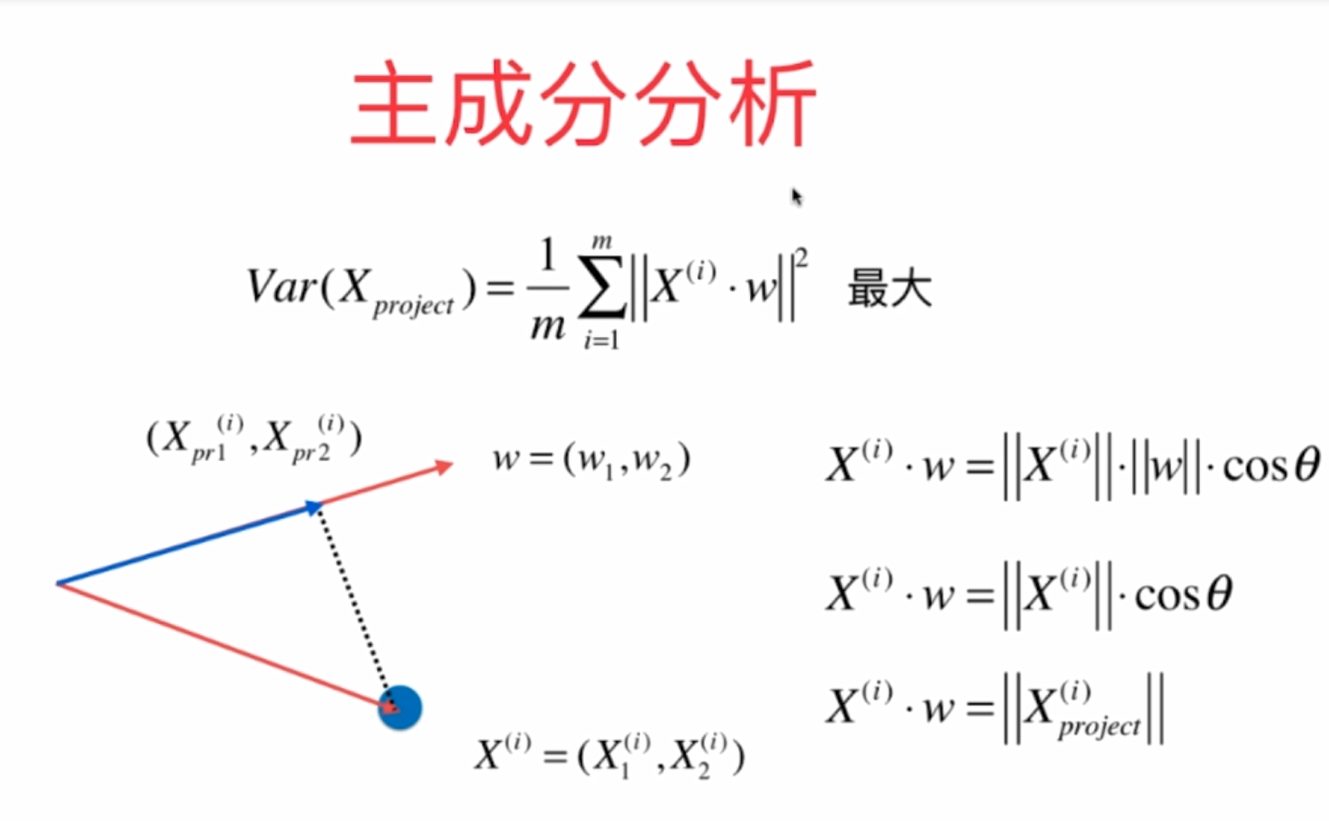

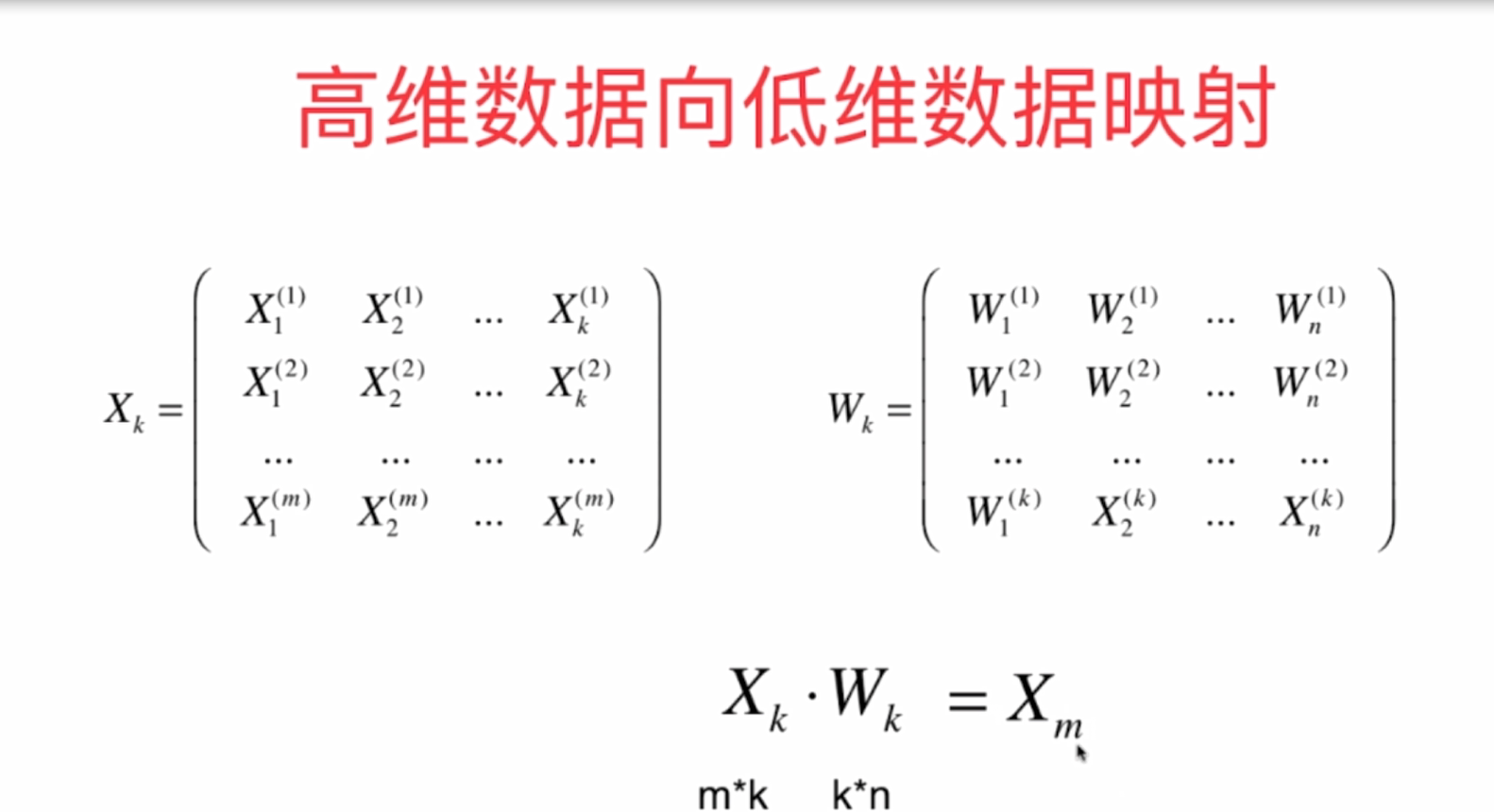
3 realize PCA algorithm by yourself
3.1 simulation of PCA by gradient rise method
1. Simulation preparation of data
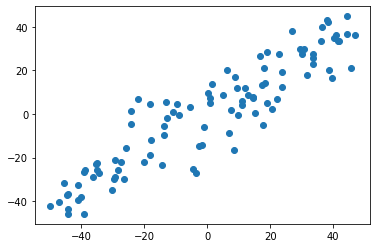
import numpy as np import matplotlib.pyplot as plt X = np.empty((100, 2)) X[:,0] = np.random.uniform(0., 100., size=100) X[:,1] = 0.75 * X[:,0] + 3. + np.random.normal(0, 10., size=100) plt.scatter(X[:,0], X[:,1]) plt.show()

2.demean operation (for each feature, set the mean value to 0 according to the column of X)
def demean(X): return X - np.mean(X, axis=0) X_demean = demean(X)
3. Use gradient rise method
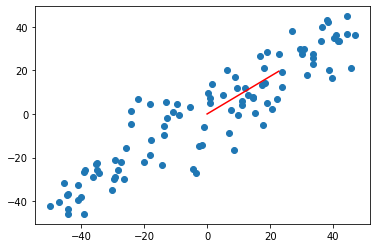
def f(w, X): return np.sum((X.dot(w)**2)) / len(X) def df_math(w, X): return X.T.dot(X.dot(w)) * 2. / len(X) def df_debug(w, X, epsilon=0.0001): res = np.empty(len(w)) for i in range(len(w)): w_1 = w.copy() w_1[i] += epsilon w_2 = w.copy() w_2[i] -= epsilon res[i] = (f(w_1, X) - f(w_2, X)) / (2 * epsilon) return res def direction(w): return w / np.linalg.norm(w) # Unit vectors def gradient_ascent(df, X, initial_w, eta, n_iters = 1e4, epsilon=1e-8): w = direction(initial_w) cur_iter = 0 while cur_iter < n_iters: gradient = df(w, X) last_w = w w = w + eta * gradient w = direction(w) # Note 1: one unit direction at a time if(abs(f(w, X) - f(last_w, X)) < epsilon): break cur_iter += 1 return w initial_w =np.random.random(X.shape[1]) # Note that 2 cannot start with the 0 vector w = gradient_ascent(df_math,X_demean,initial_w,eta) plt.scatter(X_demean[:,0],X_demean[:,1]) plt.plot([0,w[0]*30],[0,w[1]*30],color = 'r') plt.show()
Note 3: StandardScaler cannot be used to standardize data
4. The first principal component is calculated above. If other principal components are calculated, the first principal component should be subtracted and then put into it for solution.
#Other principal components X2 = np.empty(X.shape) for i in range(len(X)): X2[i] = X[i] - X[i].dot(w)*w #Remove the first principal component #Or it can be expressed as X2 = X - X.dot(w).reshape(-1,1)*w
5. Find the first n principal components
def first_n_components(n,X,eta=0.01,n_iters=1e4,epsilon=1e-8): X_pca = X.copy() X_pca = demean(X_pca) res = [] # Main ingredients for storage for i in range(n): initial_w = np.random.random(X_pca.shape[1]) w = first_component(df,X_pca,initial_w,eta) res.append(w) X_pca = X_pca - X_pca.dot(w).reshape(-1,1)*w return res first_n_components(2,X) #call
5. Realize PCA and encapsulate it
import numpy as np class PCA: def __init__(self, n_components): """Initialization PCA""" assert n_components >= 1, "n_components must be valid" self.n_components = n_components self.components_ = None def fit(self, X, eta=0.01, n_iters=1e4): """Get data set X Before n Principal components""" assert self.n_components <= X.shape[1], \ "n_components must not be greater than the feature number of X" def demean(X): return X - np.mean(X, axis=0) def f(w, X): return np.sum((X.dot(w) ** 2)) / len(X) def df(w, X): return X.T.dot(X.dot(w)) * 2. / len(X) def direction(w): return w / np.linalg.norm(w) def first_component(X, initial_w, eta=0.01, n_iters=1e4, epsilon=1e-8): w = direction(initial_w) cur_iter = 0 while cur_iter < n_iters: gradient = df(w, X) last_w = w w = w + eta * gradient w = direction(w) if (abs(f(w, X) - f(last_w, X)) < epsilon): break cur_iter += 1 return w X_pca = demean(X) self.components_ = np.empty(shape=(self.n_components, X.shape[1])) for i in range(self.n_components): initial_w = np.random.random(X_pca.shape[1]) w = first_component(X_pca, initial_w, eta, n_iters) self.components_[i,:] = w X_pca = X_pca - X_pca.dot(w).reshape(-1, 1) * w return self def transform(self, X): """Given X,Mapping to principal components""" assert X.shape[1] == self.components_.shape[1] return X.dot(self.components_.T) def inverse_transform(self, X): """Given X,Reverse mapping back to the original feature space""" assert X.shape[1] == self.components_.shape[0] return X.dot(self.components_) def __repr__(self): return "PCA(n_components=%d)" % self.n_components
- Dimension reduction of data
import numpy as np import matplotlib.pyplot as plt X = np.empty((100, 2)) X[:,0] = np.random.uniform(0., 100., size=100) X[:,1] = 0.75 * X[:,0] + 3. + np.random.normal(0, 10., size=100) from playML.PCA import PCA pca = PCA(n_components=2) pca.fit(X) pca.components_ pca = PCA(n_components=1) pca.fit(X) X_reduction = pca.transform(X) X_reduction.shape X_restore = pca.inverse_transform(X_reduction) X_restore.shape plt.scatter(X[:,0], X[:,1], color='b', alpha=0.5) plt.scatter(X_restore[:,0], X_restore[:,1], color='r', alpha=0.5) plt.show()
N? Components represents the required principal components. X ﹣ reduction is the matrix after dimension reduction. X ﹣ restore represents the matrix recovered after dimension reduction.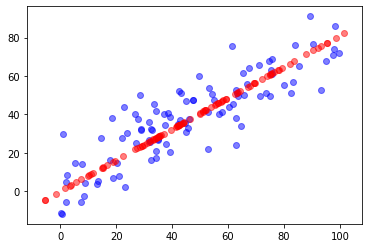
PCA algorithm in 4 sklearn
1. Reduce dimension by pca
from sklearn.decomposition import PCA pca = PCA(n_components=1) pca.fit(X) pca.components_ X_reduction = pca.transform(X) X_restore = pca.inverse_transform(X_reduction)
2. After dimension reduction by PCA with specific handwritten digit recognition, KNN is used for classification
import numpy as np import matplotlib.pyplot as plt from sklearn import datasets digits = datasets.load_digits() X = digits.data y = digits.target from sklearn.model_selection import train_test_split X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=666) from sklearn.decomposition import PCA pca = PCA(n_components=2) pca.fit(X_train) X_train_reduction = pca.transform(X_train) X_test_reduction = pca.transform(X_test) knn_clf = KNeighborsClassifier() knn_clf.fit(X_train_reduction, y_train) knn_clf.score(X_test_reduction, y_test)
3. Take the number of principal component components to explain the principal component fraction (PCA. Explained "variance" explained by principal component)
from sklearn.decomposition import PCA pca = PCA(n_components=X_train.shape[1]) pca.fit(X_train) plt.plot([i for i in range(X_train.shape[1])], [np.sum(pca.explained_variance_ratio_[:i+1]) for i in range(X_train.shape[1])]) plt.show()
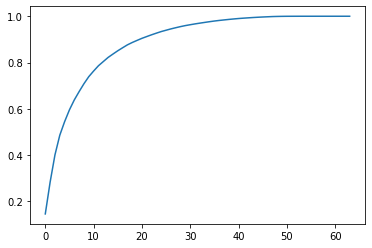
Or use the following:
pca = PCA(0.95) pca.fit(X_train) pca.n_components_ X_train_reduction = pca.transform(X_train) X_test_reduction = pca.transform(X_test) knn_clf = KNeighborsClassifier() knn_clf.fit(X_train_reduction, y_train) knn_clf.score(X_test_reduction, y_test)
PCA. N ﹣ components ﹣ is the number of all principal components whose corresponding proportion is 0.95
4. Data visualization after dimensionality reduction
%%time pca = PCA(n_components=2) pca.fit(X) X_reduction = pca.transform(X) for i in range(10): plt.scatter(X_reduction[y==i,0], X_reduction[y==i,1], alpha=0.8)
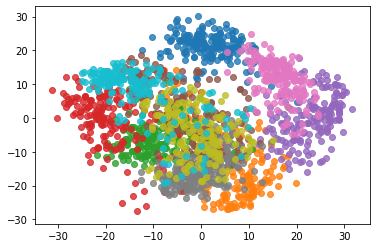
5 other applications of PCA
1. Noise reduction by PCA
from sklearn import datasets digits = datasets.load_digits() X = digits.data y = digits.target noisy_digits = X + np.random.normal(0, 4, size=X.shape) example_digits = noisy_digits[y==0,:][:10] for num in range(1,10): example_digits = np.vstack([example_digits, noisy_digits[y==num,:][:10]]) example_digits.shape def plot_digits(data): fig, axes = plt.subplots(10, 10, figsize=(10, 10), subplot_kw={'xticks':[], 'yticks':[]}, gridspec_kw=dict(hspace=0.1, wspace=0.1)) for i, ax in enumerate(axes.flat): ax.imshow(data[i].reshape(8, 8), cmap='binary', interpolation='nearest', clim=(0, 16)) plt.show() plot_digits(example_digits) pca = PCA(0.5).fit(noisy_digits) pca.n_components_ components = pca.transform(example_digits) filtered_digits = pca.inverse_transform(components) plot_digits(filtered_digits) components = pca.transform(example_digits) filtered_digits = pca.inverse_transform(components) plot_digits(filtered_digits)
Using the positive and negative transformation of PCA to remove the existing noise
2. feature faces
import numpy as np import matplotlib.pyplot as plt from sklearn.datasets import fetch_lfw_people faces = fetch_lfw_people() faces.keys() faces.data.shape faces.target_names faces.images.shape random_indexes = np.random.permutation(len(faces.data)) X = faces.data[random_indexes] example_faces = X[:36,:] example_faces.shape def plot_faces(faces): fig, axes = plt.subplots(6, 6, figsize=(10, 10), subplot_kw={'xticks':[], 'yticks':[]}, gridspec_kw=dict(hspace=0.1, wspace=0.1)) for i, ax in enumerate(axes.flat): ax.imshow(faces[i].reshape(62, 47), cmap='bone') plt.show() plot_faces(example_faces) # Characteristic faces from sklearn.decomposition import PCA pca = PCA(svd_solver='randomized') pca.fit(X) pca.components_.shape plot_faces(pca.components_[:36,:]) faces2 = fetch_lfw_people(min_faces_per_person=60) faces2.data.shape faces2.target_names len(faces2.target_names) len(faces2.target_names)
5 Summary
- When PCA is used for dimensionality reduction, it is suitable for data with more features. However, it should be noted that the data cannot be standardized, otherwise the PCA dimension reduction will fail. The basic principle is to use the gradient rise method to maximize the variance. Standardization will change the variance, so it cannot be standardized.
- PCA can also be used for noise reduction. In the process of dimension reduction, the main components are actually extracted and the noise is filtered indirectly, so the accuracy of the model can be improved.
- pca can also be used to represent feature faces.

