introduce
In this experiment, we will use support vector machine (SVM) and understand its working principle on data.
The data sets used in this experiment include:
- ex2data1.mat - linear SVM classification dataset
- ex2data2.mat - Gaussian kernel SVM classification dataset
- ex2data3.mat - cross validation Gaussian kernel SVM classification dataset
The scoring criteria are as follows:
- Point 1: use linear SVM -----------------(20 points)
- Point 2: define Gaussian kernel ---------------------(20 points)
- Point 3: use Gaussian kernel SVM ---------------(20 points)
- Key point 4: search for the optimal parameters of SVM ------------(20 points)
- Point 5: handwritten numeral recognition ---------------(20 points)
In [120]:
# Import the required library files import numpy as np import pandas as pd import matplotlib.pyplot as plt from scipy.io import loadmat import os %matplotlib inline
1 linear SVM
In this part of the experiment, linear SVM classification will be realized and applied to data set 1.
In [121]:
raw_data = loadmat('ex2data1.mat')
data = pd.DataFrame(raw_data.get('X'), columns=['X1', 'X2'])
data['y'] = raw_data.get('y')
data.head()Out[121]:
| X1 | X2 | y | |
|---|---|---|---|
| 0 | 1.9643 | 4.5957 | 1 |
| 1 | 2.2753 | 3.8589 | 1 |
| 2 | 2.9781 | 4.5651 | 1 |
| 3 | 2.9320 | 3.5519 | 1 |
| 4 | 3.5772 | 2.8560 | 1 |
In [122]:
# Define data visualization functions
def plot_init_data(data, fig, ax):
positive = data[data['y'].isin([1])]
negative = data[data['y'].isin([0])]
ax.scatter(positive['X1'], positive['X2'], s=50, marker='x', label='Positive')
ax.scatter(negative['X1'], negative['X2'], s=50, marker='o', label='Negative')In [123]:
# Data visualization fig, ax = plt.subplots(figsize=(9,6)) plot_init_data(data, fig, ax) ax.legend() plt.show()
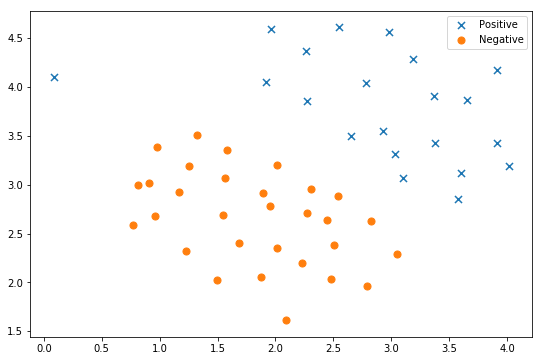
**Important point 1: * * the * * task in this part is to apply linear SVM to dataset 1: ` ex2data1.mat '** You can call the sklearn library to realize the SVM function.
In [124]:
from sklearn import svm # ======================Fill in the code here======================= svc = svm.SVC(kernel="linear") svc.fit(data[['X1', 'X2']], data['y']) svc.score(data[['X1', 'X2']], data['y']) # =============================================================
Out[124]:
0.9803921568627451
In [125]:
#Define visual classification boundary functions
def find_decision_boundary(svc, x1min, x1max, x2min, x2max, diff):
x1 = np.linspace(x1min, x1max, 1000)
x2 = np.linspace(x2min, x2max, 1000)
cordinates = [(x, y) for x in x1 for y in x2]
x_cord, y_cord = zip(*cordinates)
c_val = pd.DataFrame({'x1':x_cord, 'x2':y_cord})
c_val['cval'] = svc.decision_function(c_val[['x1', 'x2']])
decision = c_val[np.abs(c_val['cval']) < diff]
return decision.x1, decision.x2In [126]:
#Display classification decision surface
x1, x2 = find_decision_boundary(svc, 0, 4, 1.5, 5, 2 * 10**-3)
fig, ax = plt.subplots(figsize=(9,6))
ax.scatter(x1, x2, s=10, c='r',label='Boundary')
plot_init_data(data, fig, ax)
ax.set_title('SVM (C=1) Decision Boundary')
ax.legend()
plt.show()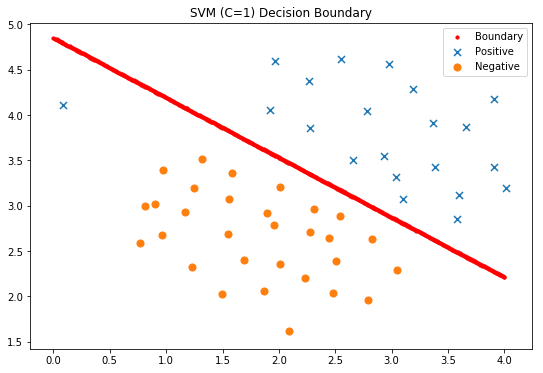
In [127]:
#Try C=100 svc2 = svm.LinearSVC(C=100, loss='hinge', max_iter=1000) svc2.fit(data[['X1', 'X2']], data['y']) svc2.score(data[['X1', 'X2']], data['y'])
/opt/conda/lib/python3.6/site-packages/sklearn/svm/base.py:931: ConvergenceWarning: Liblinear failed to converge, increase the number of iterations. "the number of iterations.", ConvergenceWarning)
Out[127]:
0.9411764705882353
In [128]:
#Display classification decision surface
x1, x2 = find_decision_boundary(svc2, 0, 4, 1.5, 5, 2 * 10**-3)
fig, ax = plt.subplots(figsize=(9,6))
ax.scatter(x1, x2, s=10, c='r',label='Boundary')
plot_init_data(data, fig, ax)
ax.set_title('SVM (C=100) Decision Boundary')
ax.legend()
plt.show()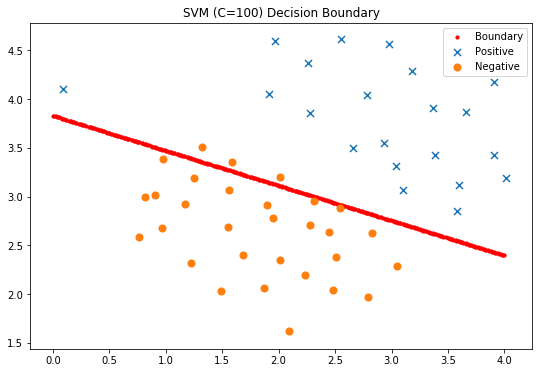
2 Gaussian kernel SVM
In this part of the experiment, the kernel SVM will be used to realize the nonlinear classification task.
2.1 Gaussian kernel
For two samples x1,x2 ∈ Rdx1,x2 ∈ Rd, the Gaussian kernel is defined as
Kgaussian(x1,x2)=exp(−∥x1−x2∥222σ2)Kgaussian(x1,x2)=exp(−‖x1−x2‖222σ2)
**Key point 2: * * the * * task of this part is to realize the definition of Gaussian kernel function according to the above formula**
In [129]:
def gaussianKernel(x1, x2, sigma):
"""
Define Gaussian kernel.
input parameter
----------
x1 : First sample point, size(d,1)Vector of
x2 : The second sample point, the size is(d,1)Vector of
sigma : Bandwidth parameters of Gaussian kernel
Output results
-------
sim : Similarity of two samples (similarity).
"""
# ======================Fill in the code here=======================
sim=np.exp(-np.power(x1-x2, 2).sum()/(2*sigma**2))
# =============================================================
return simIf the above Gaussian kernel function is completed Gaussian kernel, the following code can be used for testing. If the result is 0.324652, the calculation passes.
In [130]:
#Test Gaussian kernel function
x1 = np.array([1, 2, 1])
x2 = np.array([0, 4, -1])
sigma = 2
sim = gaussianKernel(x1, x2, sigma)
print('Gaussian Kernel between x1 and x2 is :', sim)Gaussian Kernel between x1 and x2 is : 0.32465246735834974
2.2 Gaussian kernel SVM applied to dataset 2
In this experiment, Gaussian kernel SVM is applied to data set 2: ex2data2.mat..
In [131]:
raw_data = loadmat('ex2data2.mat')
data = pd.DataFrame(raw_data['X'], columns=['X1', 'X2'])
data['y'] = raw_data['y']
fig, ax = plt.subplots(figsize=(9,6))
plot_init_data(data, fig, ax)
ax.legend()
plt.show()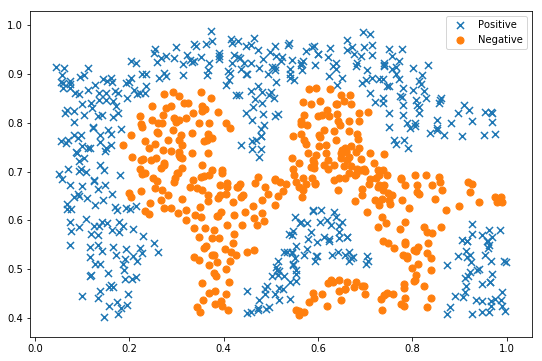
As can be seen from the above figure, the above two types of samples are linearly inseparable. Kernel SVM is needed for classification.
**Point 3: * * the * * task of this part is to apply Gaussian kernel SVM to dataset 2** The sklearn library can be called to realize the nonlinear SVM function.
In [132]:
# ======================Fill in the code here======================= svc = svm.SVC( kernel='rbf', gamma=30) svc.fit(data[['X1', 'X2']], data['y']) svc.score(data[['X1', 'X2']], data['y']) # =============================================================
Out[132]:
0.9721900347624566
In [133]:
x1, x2 = find_decision_boundary(svc, 0, 1, 0.4, 1, 0.01) fig, ax = plt.subplots(figsize=(9,6)) plot_init_data(data, fig, ax) ax.scatter(x1, x2, s=10) plt.show()
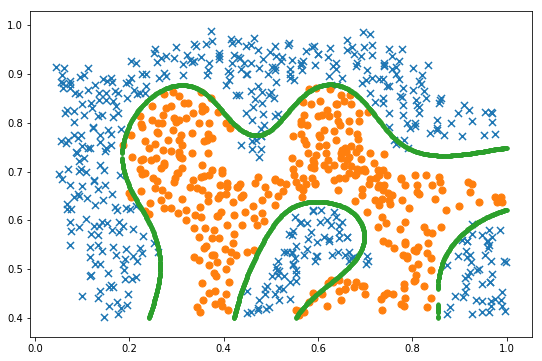
3 cross validation Gaussian kernel SVM
In this part of the experiment, the optimal parameters CC and CC of Gaussian kernel SVM will be selected by cross validation method σσ, And apply it to dataset 3: ex2data3.mat. The data set contains training sample set X (training sample characteristics), y (training sample marker) and validation set Xval (validation sample characteristics), yval (validation sample mark).
In [134]:
raw_data = loadmat('ex2data3.mat')
X = raw_data['X']
Xval = raw_data['Xval']
y = raw_data['y'].ravel()
yval = raw_data['yval'].ravel()
fig, ax = plt.subplots(figsize=(9,6))
data = pd.DataFrame(raw_data.get('X'), columns=['X1', 'X2'])
data['y'] = raw_data.get('y')
plot_init_data(data, fig, ax)
plt.show()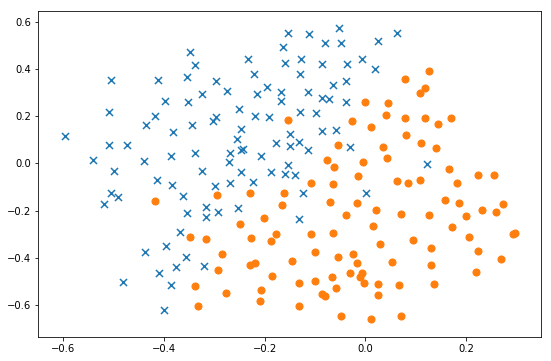
3.1 search SVM optimal parameters CC and σσ
**Key point 4: * * the * * task of this part is to search the optimal parameters CC and CC of SVM σσ. ** For CC and σσ, You can search from the following candidate sets {0.01,0.03,0.1,0.3,1,3,10,30}{0.01,0.03,0.1,0.3,1,3,10,30}
In [135]:
C_values = [0.01, 0.03, 0.1, 0.3, 1, 3, 10, 30, 100]
gamma_values = [0.01, 0.03, 0.1, 0.3, 1, 3, 10, 30, 100]
best_score = 0
# ======================Fill in the code here=======================
dataval = pd.DataFrame(raw_data.get('Xval'), columns=['X1', 'X2'])
dataval['y'] = raw_data.get('yval')
for c in C_values:
for gamma in gamma_values:
svc = svm.SVC(C=c, gamma=gamma)
svc.fit(data[['X1', 'X2']], data['y'])
score =svc.score(dataval[['X1', 'X2']], dataval['y'])
if score>best_score:
best_score=score
best_C=c
best_gamma=gamma
# =============================================================
best_C, best_gamma, best_scoreOut[135]:
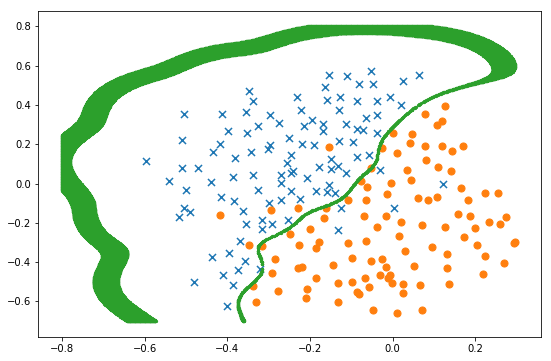
(0.3, 100, 0.965)
3.2 using the selected parameters and Gaussian kernel SVM applied to the data set 3
In [136]:
svc = svm.SVC(C=best_C, gamma=best_gamma) svc.fit(X, y) x1, x2 = find_decision_boundary(svc, -0.8, 0.3, -0.7, 0.8, 0.005) fig, ax = plt.subplots(figsize=(9,6)) plot_init_data(data, fig, ax) ax.scatter(x1, x2, s=5) plt.show()
4 apply SVM to handwritten numeral recognition
In this part of the experiment, linear SVM and Gaussian kernel SVM are applied to handwritten dataset: UCI ml hand written digits datasets, and the recognition results are compared.
In [137]:
# Import the required library files from sklearn import datasets, svm, metrics from sklearn.model_selection import train_test_split
In [138]:
# Download the dataset from the sklearn library and show some samples
digits = datasets.load_digits()
_, axes = plt.subplots(1, 10)
images_and_labels = list(zip(digits.images, digits.target))
for ax, (image, label) in zip(axes, images_and_labels[0:10]):
ax.set_axis_off()
ax.imshow(image, cmap=plt.cm.gray_r, interpolation='nearest')
ax.set_title(' %i' % label)
plt.show()
In [139]:
Turn each picture sample into a vector
n_samples = len(digits.images)
data = digits.images.reshape((n_samples, -1))
# Divide the original data set into training set and test set (half training and the other half testing)
X_train, X_test, y_train, y_test = train_test_split(
data, digits.target, test_size=0.5, shuffle=False)#False**Key point 5: * * the task of this part is * * to apply linear SVM (C=1) and Gaussian kernel SVM (C=1, gamma=0.001) to UCI handwritten dataset and output recognition accuracy**
In [140]:
#The linear SVM is applied to the data set and the recognition results are output
# ======================Fill in the code here=======================
svc = svm.SVC(kernel="linear")
svc.fit(X_train, y_train)
score_Linear=svc.score(X_test, y_test)
# =============================================================
print("Classification accuracy of Linear SVM:", score_Linear) Classification accuracy of Linear SVM: 0.9443826473859844
In [141]:
#The linear SVM is applied to the data set and the recognition results are output
# ======================Fill in the code here=======================
svc = svm.SVC(kernel="linear")
svc.fit(X_train, y_train)
score_Linear=svc.score(X_test, y_test)
# =============================================================
print("Classification accuracy of Linear SVM:", score_Linear) Classification accuracy of Gaussian SVM: 0.9688542825361512