In medical image processing, we often use a NIFTI format image (. nii file). Now let's take a look
1 NIFTI format image
1.1 what is a NIFTI format image
Before explaining what is the NIFTI (neuroimaging information technology initiative) format, you must first understand the Analyze format. Each group of data group stored in Analyze format contains two files, one is a data file with. img extension, including binary image data; The other is a header file with an extension of. hdr, which contains the metadata of the image. In the early days of FMRI, Analyze format was the most commonly used format, but now it is gradually replaced by NIFTI format. The main disadvantage of Analyze format is that the header file can not really reflect metadata.
The extension of the standard NIFTI image is. nii, which also contains header files and image data. Due to the relationship between NIFTI format and Analyze format, NIFTI format can also use independent image file (. img) and header file (. hdr). The advantage of a separate. nii format file is that it can be compressed using standard compression software (such as gzip), and some analysis software packages (such as FSL) can directly read and write the compressed. nii file (extension. nii.gz).
1.2 why does NIFTI format image appear
The original ANALYZE 7.5 format image format lacks some information, such as no direction information, left and right directions of the patient, etc. if additional information needs to be included, an additional file is required. For example, ANALYZE 7.5 needs a pair of <. HDR,. Img > files to save the complete information of the image. Therefore, to solve this problem, the Data Format Working Group (DFWG) completely defines the image format as NIFTI format.
For more information about NIFTI format, see: https://brainder.org/2012/09/23/the-nifti-file-format/
2 read NIFTI format image
2.1 ITK-SNAP
Recommended ITK-SNAP download address: http://www.itksnap.org/pmwiki/pmwiki.php?n=Downloads.SNAP3
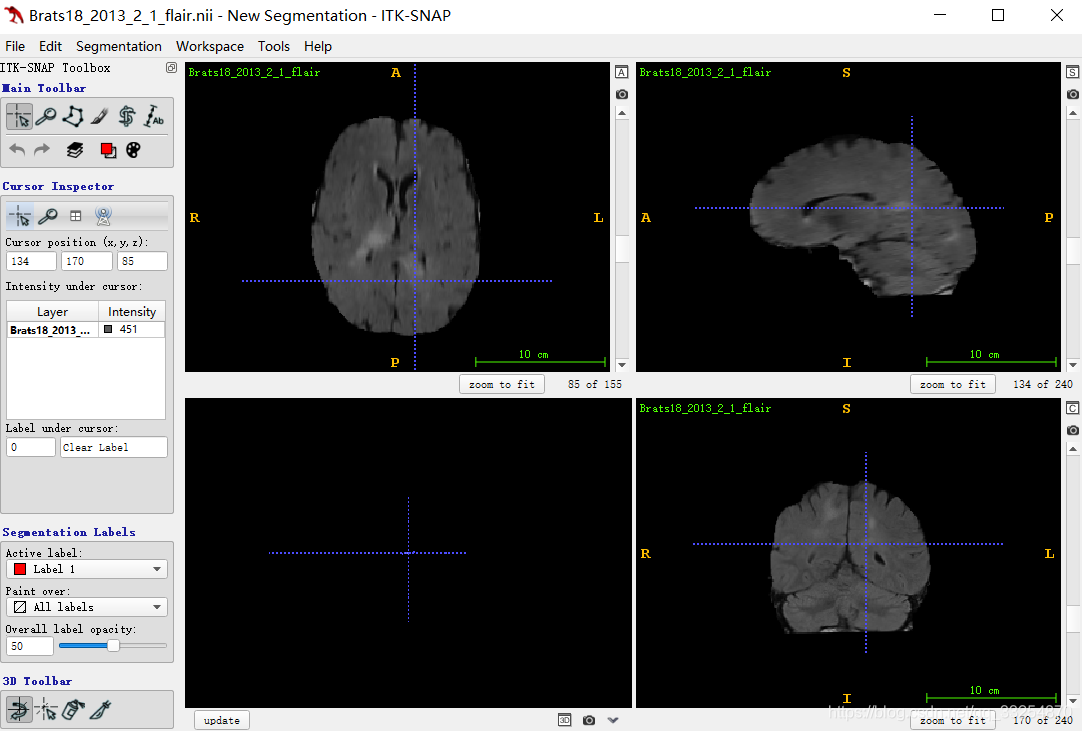
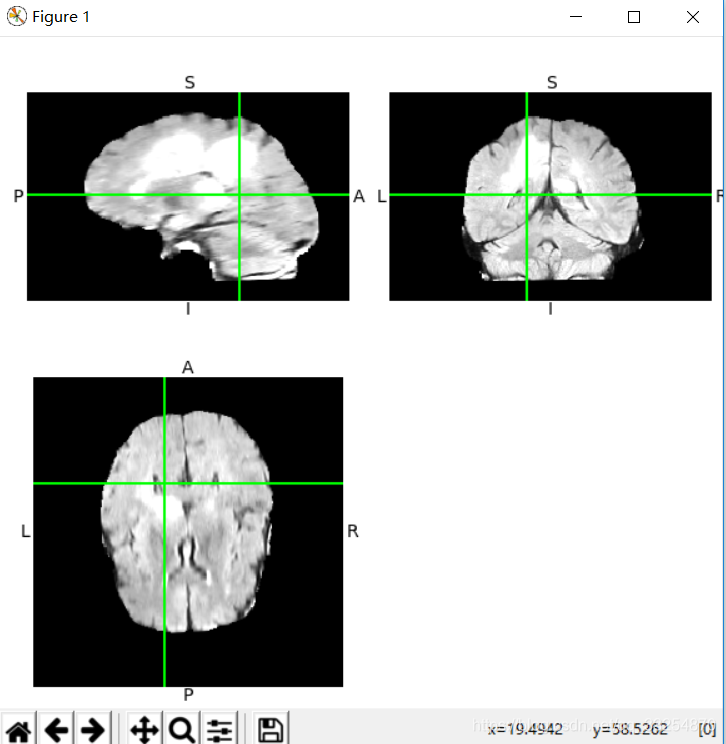
It can be seen that ITK snap obtains views from three angles (horizontal plane, sagittal plane and coronal plane)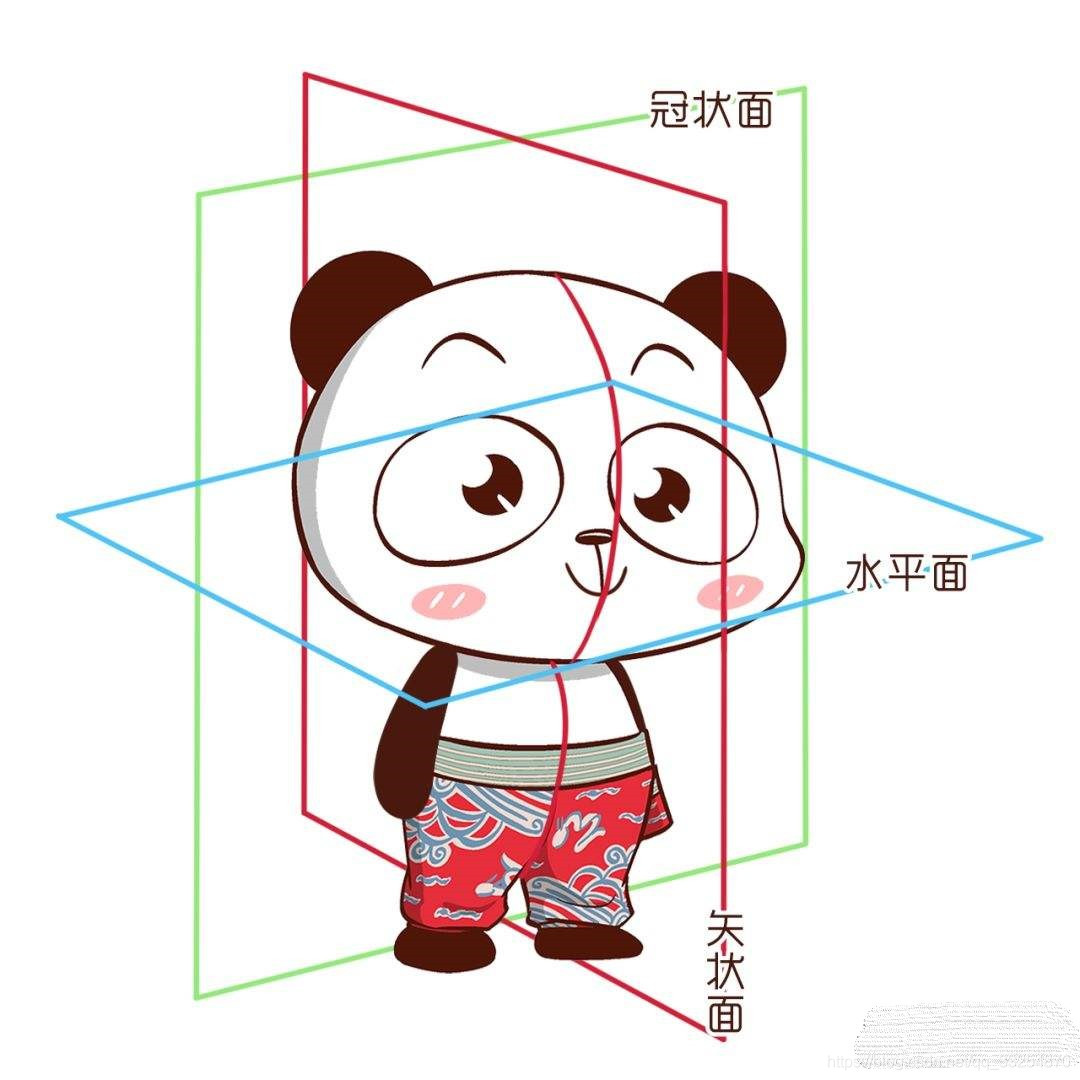
2.2 itkwidgets
For details, see: https://pypi.org/project/itkwidgets/
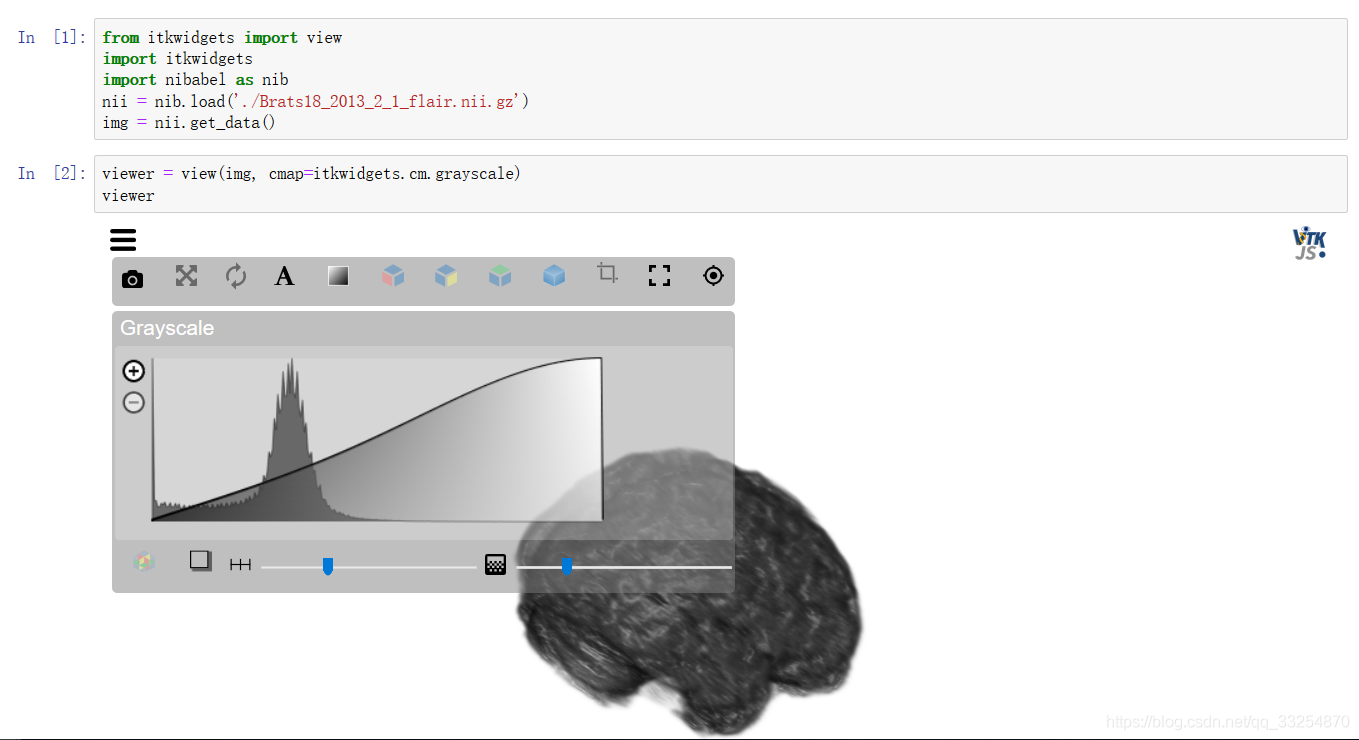
2.3 simpleITK
This is the most used image processing tool
import SimpleITK as sitk
from matplotlib import pyplot as plt
def showNii(img):
for i in range(img.shape[0]):
print(i)
plt.imshow(img[i, :, :], cmap='gray')
plt.show()
itk_img = sitk.ReadImage('./data/00_img.nii.gz')
img = sitk.GetArrayFromImage(itk_img)
print(img.shape) # (88, 132, 175) indicates the number of slices in each dimension
showNii(img)
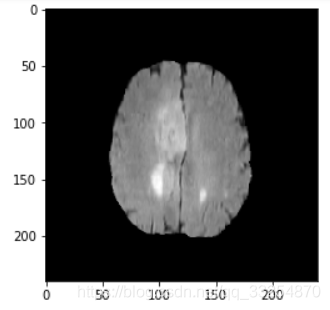
2.4 Nibabel
import matplotlib
matplotlib.use('TKAgg')
from matplotlib import pylab as plt
import nibabel as nib
from nibabel import nifti1
from nibabel.viewers import OrthoSlicer3D
example_filename = './data/00_img.nii.gz'
img = nib.load(example_filename)
print('img:\n', img)
print('img.header:\n', img.header['db_name']) # Output header information
# It is determined by the dimension of the file itself, which may be 3D or 4D
width, height, queue = img.dataobj.shape
print(width, height, queue) # 175 132 88
OrthoSlicer3D(img.dataobj).show()
num = 1
for i in range(0, queue, 11):
img_arr = img.dataobj[:, :, i]
plt.subplot(2, 4, num)
plt.imshow(img_arr, cmap='gray')
num += 1
plt.show()
reference
- https://blog.csdn.net/qq_33254870/article/details/100125788
- https://blog.csdn.net/alxe_made/article/details/80512423