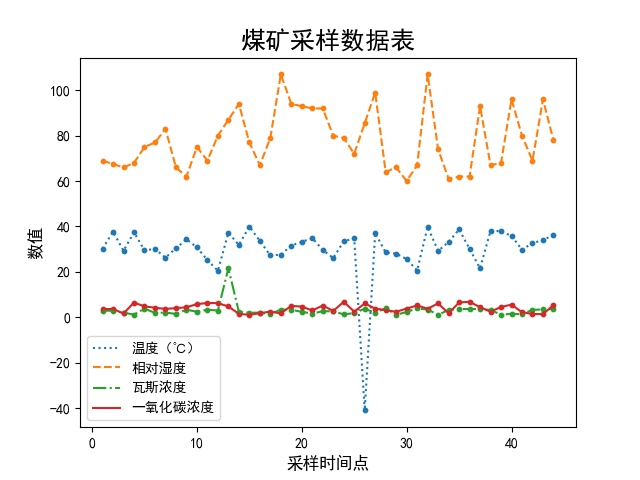
preface
In order to cope with my homework, I wanted to find a code on the Internet and handed it in. I found that I didn't have what I wanted, so I wrote it in tears for two days. In order to study hard, I rolled it up with my classmates 🤣
If you need to hand in your homework, just look at the source code directly. In the CleaningData () method, you must change it to the data you need. I've written everything else for you
target
Deal with missing and abnormal values in data (find out and re assign values)
process
Blank value (missing value)
For the processing of blank value, I use the average value of the two lines before and after the blank value to assign a value;
If the first and second lines are blank values, the position will always be found;
If a column has only one data, I will use this data to assign a value to the whole column ψ (`∇´) ψ
(at least I hope the last situation doesn't happen)
Outliers
Exception handling, I use Three point Lagrange interpolation
Why use this? Because the teacher asked ƪ ( ˘ ⌣ ˘)ʃ
Use three points to find an unknown equation, and use this equation to predict the value of abnormal value position.
This equation can be worked out. You can try to output it. (shown below)
harvest
Harvest 1:
# Show all columns
pd.set_option('display.max_columns', None)
# Show all rows
pd.set_option('display.max_rows', None)
# Align column names
pd.set_option('display.unicode.ambiguous_as_wide', True)
pd.set_option('display.unicode.east_asian_width', True)
# Cancel scientific counting output
np.set_printoptions(suppress=True)
# In order to display the numbers intuitively, the scientific counting method is not used
pd.set_option('display.float_format', lambda x: '%.2f' % x)
# Used to display Chinese labels normally
plt.rcParams['font.sans-serif'] = ['SimHei']
# Used to display negative signs normally
plt.rcParams['axes.unicode_minus'] = False
Harvest 2:
np.where (data = = '1'): find the position of 1
print(np.where(data == '')) # Find the coordinates of the blank value
Harvest 3:
A valid multidimensional iterator object that can traverse an array.
Here I convert string data into floating-point numbers. Two can be implemented
np.nditer(row) or [float(num) for num in arr[:, 0]]
Harvest 4:
Three point Lagrange interpolation
from scipy.interpolate import lagrange iloc_point_of_time = [float(num) for num in arr[:, 0]][4:7:1] iloc_temperature = [float(num) for num in arr[:, 1]][4:7:1] a1 = lagrange(iloc_point_of_time, iloc_temperature) print(a1)
Harvest 5:
If you want to make the points on the line chart highlight, you only need to use plot () and scatter () at the same time
It is the combination of line chart and scatter chart
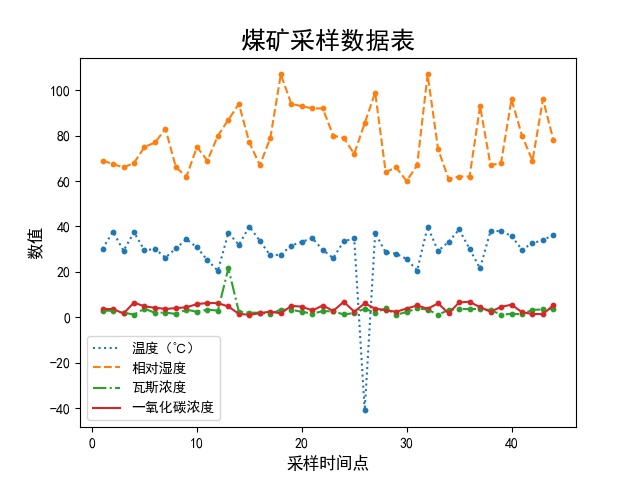
Core source code
Process blank values
Get the coordinate array of the blank array, and pass the elements of the array to row and col in fill_value (arrow, row, Col)
def Create_Fill_Value_Arr(arr): # If you need coordinates, the number of abscissa and ordinate should be the same. Create a 2-dimensional zero array with len (row) length
# row and col are sets of abscissa and ordinate, respectively
# print(np.where(data == '')) # Find the coordinates of the blank value
row = np.where(arr == '')[0]
col = np.where(arr == '')[1]
CoordinateArray = np.zeros((len(row), 2)) # Blank value coordinate array
row_count = 0
for i in np.nditer(row):
CoordinateArray[row_count][0] = i
row_count += 1
col_count = 0
for i in np.nditer(col):
CoordinateArray[col_count][1] = i
col_count += 1
return CoordinateArray
arr: array to be processed
row: abscissa of the element to be processed
col: ordinate of the element to be processed
def Fill_Value(arr, row, col):
# The two-dimensional array is filled with blank values
# There is a precondition here. This assignment is based on the upstream and downstream values of the same column, so the column must not 1 change, only the number of rows
current_row = row # Record the number of lines entered
current_col = col # Record the number of columns entered
next_row = 0 # Record the number of lines in the next line
next_col = col # Record the number of columns in the next column
pre_row = 0 # Record the number of lines in the previous line
pre_col = col # Record the number of columns in the previous column
# Assignment of the first and last lines
if row == 0 or row == len(arr) - 1:
if row == 0: # If the position of the input element is in the first row, the non empty number of the next row in the same column is assigned to it
flag = True # Tag, if the assignment ends, the loop ends
while flag:
current_row += 1
if arr[current_row][current_col] != '':
arr[row][col] = arr[current_row][current_col] # assignment
flag = False
elif arr[current_row][current_col] == '': # Judge if the current number is null
current_row += 1 # Then find the value of the next row in the same column
else:
print("Fill_Value()method==first line==Something went wrong!!")
elif row == len(arr) - 1: # If the position of the input element is in the last row, the non empty number of the row on the same column is assigned to it
flag = True # Tag, if the assignment ends, the loop ends
current_row -= 1
while flag:
if arr[current_row][current_col] != '':
arr[row][col] = arr[current_row][current_col]
flag = False
elif arr[current_row][current_col] == '':
current_row -= 1
else:
print("Fill_Value()Methodical==Last line==Something went wrong!!")
else:
print("If this occurs, the condition judgment of the first and last lines is wrong")
# Assignment of non first and last lines
elif row != 0 and row != len(arr) - 1: # Assign the average value of the upper and lower rows of the same column of the value to it
# There is an arithmetic pit here, that is, when you get to this line, the previous numbers must be non empty values, because the above algorithm fills the previous values,
# Therefore, for this number, the first n lines have values, but the latter ones do not necessarily have values. It is possible that many lines have null values,
# Therefore, we need to make some judgment when selecting the next value
# The practice here is to find a value to average no matter how many lines are separated
# If all rows in this row are blank values, the algorithm here assigns the previous value of this row to all subsequent rows
flag = True # Tag, if the assignment ends, the loop ends
next_row = current_row + 1
pre_row = current_row - 1
while flag:
if arr[next_row][next_col] != '': # Here we judge whether the next line is null
arr[row][col] = str((float(arr[next_row][next_col]) + # Since the numerical calculation of two numbers is involved here, it needs to be converted to numerical type, while the above only involves the assignment behavior of a single line
float(arr[pre_row][pre_col])) / 2) # Floating point numbers are required here, not integers
flag = False
elif arr[next_row][next_col] == '':
next_row += 1
else:
print("Fill_Value()Methodical==Assignment of non first and last lines==Something went wrong!!")
else:
print('How could this be wrong?')
return arr
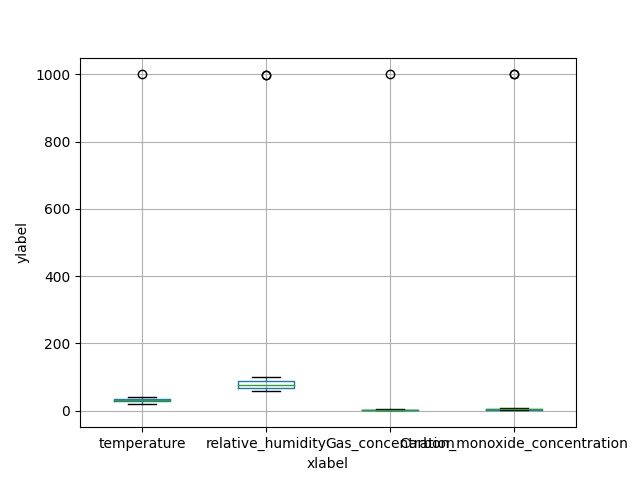
Handling outliers
You need to modify some data here, because when you write it, you find that if you keep improving, it won't be enough for me to write in a week, so I can only be lazy secretly. Forgive me (´▽ ʃ ♡ ƪ) (≧∇≦)ノ(❁´◡❁)
arr: enter the array after processing the blank value
# Abnormal data can be obtained through the box diagram
def CleaningData(arr):
# The data of arr is all strings
# If a data is very different from all the data in the table, it is necessary to replace the normal data with the abnormal data
point_of_time = [float(num) for num in arr[:, 0]] # This sampling time point should not be used
temperature = [float(num) for num in arr[:, 1]]
relative_humidity = [float(num) for num in arr[:, 2]]
Gas_concentration = [float(num) for num in arr[:, 3]]
Carbon_monoxide_concentration = [float(num) for num in arr[:, 4]]
# The data of data is all float
data = {
'temperature': temperature,
'relative_humidity': relative_humidity,
'Gas_concentration': Gas_concentration,
'Carbon_monoxide_concentration': Carbon_monoxide_concentration,
}
df = pd.DataFrame(data)
df.boxplot() # Here, pandas has its own process
plt.ylabel("ylabel")
plt.xlabel("xlabel") # We set the title of the abscissa and ordinate.
plt.savefig('Box diagram.jpg')
plt.show()
# The des data here comes from df.describe (), because
des = {
'temperature': {
'count': 44.00,
'mean': 53.63,
'std': 146.07,
'min': 20.50,
'25%': 29.06,
'50%': 31.73,
'75%': 35.93,
'max': 999.99,
},
'relative_humidity': {
'count': 44.00,
'mean': 118.39,
'std': 194.72,
'min': 60.00,
'25%': 67.00,
'50%': 76.00,
'75%': 88.25,
'max': 999.99,
},
'Gas_concentration': {
'count': 44.00,
'mean': 25.23,
'std': 150.37,
'min': 1.08,
'25%': 1.90,
'50%': 2.65,
'75%': 3.39,
'max': 999.99,
},
'Carbon_monoxide_concentration': {
'count': 44.00,
'mean': 49.15,
'std': 209.90,
'min': 1.08,
'25%': 2.47,
'50%': 3.94,
'75%': 5.23,
'max': 999.99,
}
}
columns_list = ['temperature', 'relative_humidity', 'Gas_concentration', 'Carbon_monoxide_concentration']
# If any value exceeds the average value, the value is regarded as an abnormal value
arrays_list = [] # This list is used to record the coordinates of outliers
col = 0
row = 0
for col_list in columns_list:
for i in range(len(arr)):
if data[col_list][row] >= des[col_list]['std']:
arrays_list.append([row, col])
row += 1
elif data[col_list][row] < des[col_list]['std']:
row += 1
else:
print("This can't happen once")
col += 1
row = 0
# Three values select (4, 5, 6) rows
iloc_point_of_time = [float(num) for num in arr[:, 0]][4:7:1] # This sample collection time
iloc_temperature = [float(num) for num in arr[:, 1]][4:7:1]
iloc_relative_humidity = [float(num) for num in arr[:, 2]][4:7:1]
iloc_Gas_concentration = [float(num) for num in arr[:, 3]][4:7:1]
iloc_Carbon_monoxide_concentration = [float(num) for num in arr[:, 4]][4:7:1]
# Here, the lagrange three value interpolation method is used to call the lagrange () function
a1 = lagrange(iloc_point_of_time, iloc_temperature)
a2 = lagrange(iloc_point_of_time, iloc_relative_humidity)
a3 = lagrange(iloc_point_of_time, iloc_Gas_concentration)
a4 = lagrange(iloc_point_of_time, iloc_Carbon_monoxide_concentration)
# Assign outliers
for i in range(len(arrays_list)):
if arrays_list[i][1] == 0:
arr[arrays_list[i][0]][1] = a1(arrays_list[i][1])
elif arrays_list[i][1] == 1:
arr[arrays_list[i][0]][2] = a2(arrays_list[i][1])
elif arrays_list[i][1] == 2:
arr[arrays_list[i][0]][3] = a3(arrays_list[i][1])
elif arrays_list[i][1] == 3:
arr[arrays_list[i][0]][4] = a4(arrays_list[i][1])
print(arr)
return arr
Write at the end
Because the quality of the code you write is not high, it will be a headache for you to read the whole blog.
If you really need data processing code, I'll try to extract what I think is the key content.
I hope this blog can help you and solve your problems.
Wait a little further to modify the source code.
source file
data
I think you should be able to input these data into the csv file. It's good to read them anyway
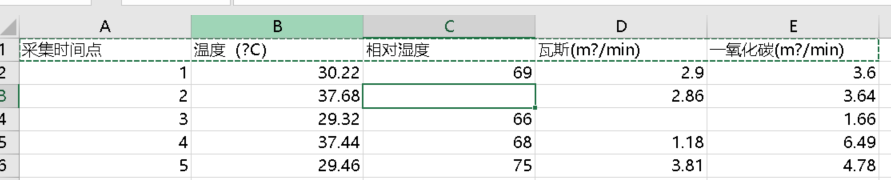 (~ ̄▽ ̄)~
(~ ̄▽ ̄)~
1,30.22,69,2.9,3.6 2,37.68,,2.86,3.64 3,29.32,66,,1.66 4,37.44,68,1.18,6.49 5,29.46,75,3.81,4.78 6,30.12,77,1.93,4.2 7,26.3,83,2.07,3.67 8,,66,1.46, 9,34.38,62,3.36,4.39 10,30.79,75,2.4,5.79 11,25.17,69,3.34,6.28 12,20.5,80,2.95,6.22 13,37.21,,999.99,4.75 14,31.88,94,2.41,1.49 15,39.94,77,1.98,1.08 16,33.65,67,2.03, 17,27.21,79,1.41,2.5 18,27.57,999,3.39,1.7 19,31.59,94,3.27,5.02 20,,93,,4.69 21,34.88,92,1.58,3.01 22,29.65,92,2.69,5.16 23,26.05,80,2.61,2.8 24,33.5,79,1.26,6.81 25,34.71,72,1.82,2.49 26,999.99,,3.77,999.99 27,37.06,99,1.94,3.85 28,28.57,64,3.93, 29,28,66,1.08,2.4 30,25.67,60,2.33,3.84 31,20.59,67,3.88,5.19 32,39.84,999,3.4,3.77 33,29.22,74,1.09,999.99 34,33.19,61,3.22,1.74 35,38.7,62,3.61,6.52 36,,62,,6.83 37,21.56,93,3.63,4.43 38,38.06,67,3.21,2.41 39,37.95,68,1.12,4.56 40,35.8,96,1.52,5.54 41,29.75,80,1.3,2.23 42,32.69,69,3.26,1.43 43,33.94,96,3.42,1.4 44,36.31,78,3.69,5.34
Source code
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from scipy.interpolate import lagrange
# Show all columns
pd.set_option('display.max_columns', None)
# Show all rows
pd.set_option('display.max_rows', None)
# Align column names
pd.set_option('display.unicode.ambiguous_as_wide', True)
pd.set_option('display.unicode.east_asian_width', True)
# Cancel scientific counting output
np.set_printoptions(suppress=True)
# In order to display the numbers intuitively, the scientific counting method is not used
pd.set_option('display.float_format', lambda x: '%.2f' % x)
'''
param :
arr: target array
row: that 's ok
col: column
'''
def Fill_Value(arr, row, col):
# The two-dimensional array is filled with blank values
# There is a precondition here. This assignment is based on the upstream and downstream values of the same column, so the column must not 1 change, only the number of rows
current_row = row # Record the number of lines entered
current_col = col # Record the number of columns entered
next_row = 0 # Record the number of lines in the next line
next_col = col # Record the number of columns in the next column
pre_row = 0 # Record the number of lines in the previous line
pre_col = col # Record the number of columns in the previous column
# Assignment of the first and last lines
if row == 0 or row == len(arr) - 1:
if row == 0: # If the position of the input element is in the first row, the non empty number of the next row in the same column is assigned to it
flag = True # Tag, if the assignment ends, the loop ends
while flag:
current_row += 1
if arr[current_row][current_col] != '':
arr[row][col] = arr[current_row][current_col] # assignment
flag = False
elif arr[current_row][current_col] == '': # Judge if the current number is null
current_row += 1 # Then find the value of the next row in the same column
else:
print("Fill_Value()method==first line==Something went wrong!!")
elif row == len(arr) - 1: # If the position of the input element is in the last row, the non empty number of the row on the same column is assigned to it
flag = True # Tag, if the assignment ends, the loop ends
current_row -= 1
while flag:
if arr[current_row][current_col] != '':
arr[row][col] = arr[current_row][current_col]
flag = False
elif arr[current_row][current_col] == '':
current_row -= 1
else:
print("Fill_Value()Methodical==Last line==Something went wrong!!")
else:
print("If this occurs, the condition judgment of the first and last lines is wrong")
# Assignment of non first and last lines
elif row != 0 and row != len(arr) - 1: # Assign the average value of the upper and lower rows of the same column of the value to it
# There is an arithmetic pit here, that is, when you get to this line, the previous numbers must be non empty values, because the above algorithm fills the previous values,
# Therefore, for this number, the first n lines have values, but the latter ones do not necessarily have values. It is possible that many lines have null values,
# Therefore, we need to make some judgment when selecting the next value
# The practice here is to find a value to average no matter how many lines are separated
# If all rows in this row are blank values, the algorithm here assigns the previous value of this row to all subsequent rows
flag = True # Tag, if the assignment ends, the loop ends
next_row = current_row + 1
pre_row = current_row - 1
while flag:
if arr[next_row][next_col] != '': # Here we judge whether the next line is null
arr[row][col] = str((float(arr[next_row][next_col]) + # Since the numerical calculation of two numbers is involved here, it needs to be converted to numerical type, while the above only involves the assignment behavior of a single line
float(arr[pre_row][pre_col])) / 2) # Floating point numbers are required here, not integers
flag = False
elif arr[next_row][next_col] == '':
next_row += 1
else:
print("Fill_Value()Methodical==Assignment of non first and last lines==Something went wrong!!")
else:
print('How could this be wrong?')
return arr
def Create_Fill_Value_Arr(arr): # If you need coordinates, the number of abscissa and ordinate should be the same. Create a 2-dimensional zero array with len (row) length
# row and col are sets of abscissa and ordinate, respectively
# print(np.where(data == '')) # Find the coordinates of the blank value
row = np.where(arr == '')[0]
col = np.where(arr == '')[1]
CoordinateArray = np.zeros((len(row), 2)) # Blank value coordinate array
row_count = 0
for i in np.nditer(row):
CoordinateArray[row_count][0] = i
row_count += 1
col_count = 0
for i in np.nditer(col):
CoordinateArray[col_count][1] = i
col_count += 1
return CoordinateArray
# Abnormal data can be obtained through the box diagram
def CleaningData(arr):
# The data of arr is all strings
# If a data is very different from all the data in the table, it is necessary to replace the normal data with the abnormal data
point_of_time = [float(num) for num in arr[:, 0]] # This sampling time point should not be used
temperature = [float(num) for num in arr[:, 1]]
relative_humidity = [float(num) for num in arr[:, 2]]
Gas_concentration = [float(num) for num in arr[:, 3]]
Carbon_monoxide_concentration = [float(num) for num in arr[:, 4]]
# The data of data is all float
data = {
'temperature': temperature,
'relative_humidity': relative_humidity,
'Gas_concentration': Gas_concentration,
'Carbon_monoxide_concentration': Carbon_monoxide_concentration,
}
df = pd.DataFrame(data)
df.boxplot() # Here, pandas has its own process
plt.ylabel("ylabel")
plt.xlabel("xlabel") # We set the title of the abscissa and ordinate.
plt.savefig('Box diagram.jpg')
plt.show()
# The des data here comes from df.describe (), because
des = {
'temperature': {
'count': 44.00,
'mean': 53.63,
'std': 146.07,
'min': 20.50,
'25%': 29.06,
'50%': 31.73,
'75%': 35.93,
'max': 999.99,
},
'relative_humidity': {
'count': 44.00,
'mean': 118.39,
'std': 194.72,
'min': 60.00,
'25%': 67.00,
'50%': 76.00,
'75%': 88.25,
'max': 999.99,
},
'Gas_concentration': {
'count': 44.00,
'mean': 25.23,
'std': 150.37,
'min': 1.08,
'25%': 1.90,
'50%': 2.65,
'75%': 3.39,
'max': 999.99,
},
'Carbon_monoxide_concentration': {
'count': 44.00,
'mean': 49.15,
'std': 209.90,
'min': 1.08,
'25%': 2.47,
'50%': 3.94,
'75%': 5.23,
'max': 999.99,
}
}
columns_list = ['temperature', 'relative_humidity', 'Gas_concentration', 'Carbon_monoxide_concentration']
# If any value exceeds the average value, the value is regarded as an abnormal value
arrays_list = [] # This list is used to record the coordinates of outliers
col = 0
row = 0
for col_list in columns_list:
for i in range(len(arr)):
if data[col_list][row] >= des[col_list]['std']:
arrays_list.append([row, col])
row += 1
elif data[col_list][row] < des[col_list]['std']:
row += 1
else:
print("This can't happen once")
col += 1
row = 0
# Three values select (4, 5, 6) rows
iloc_point_of_time = [float(num) for num in arr[:, 0]][4:7:1] # This sample collection time
iloc_temperature = [float(num) for num in arr[:, 1]][4:7:1]
iloc_relative_humidity = [float(num) for num in arr[:, 2]][4:7:1]
iloc_Gas_concentration = [float(num) for num in arr[:, 3]][4:7:1]
iloc_Carbon_monoxide_concentration = [float(num) for num in arr[:, 4]][4:7:1]
# Here, the lagrange three value interpolation method is used to call the lagrange () function
a1 = lagrange(iloc_point_of_time, iloc_temperature)
a2 = lagrange(iloc_point_of_time, iloc_relative_humidity)
a3 = lagrange(iloc_point_of_time, iloc_Gas_concentration)
a4 = lagrange(iloc_point_of_time, iloc_Carbon_monoxide_concentration)
# Assign outliers
for i in range(len(arrays_list)):
if arrays_list[i][1] == 0:
arr[arrays_list[i][0]][1] = a1(arrays_list[i][1])
elif arrays_list[i][1] == 1:
arr[arrays_list[i][0]][2] = a2(arrays_list[i][1])
elif arrays_list[i][1] == 2:
arr[arrays_list[i][0]][3] = a3(arrays_list[i][1])
elif arrays_list[i][1] == 3:
arr[arrays_list[i][0]][4] = a4(arrays_list[i][1])
# print(arr)
return arr
# Represent the data in the form of images
def CreatePicture(arr):
# Here, corresponding variables are created for files, which may not be applicable to other files
# Because the initial value is a string, you need to convert the string in the list to floating point, otherwise an error will be reported
point_of_time = [float(num) for num in arr[:, 0]]
temperature = [float(num) for num in arr[:, 1]]
relative_humidity = [float(num) for num in arr[:, 2]]
Gas_concentration = [float(num) for num in arr[:, 3]]
Carbon_monoxide_concentration = [float(num) for num in arr[:, 4]]
# Connect each point into a line segment
plt.plot(point_of_time, temperature, label="Temperature(℃)", linestyle=":")
plt.plot(point_of_time, relative_humidity, label="relative humidity", linestyle="--")
plt.plot(point_of_time, Gas_concentration, label="Gas concentration", linestyle="-.")
plt.plot(point_of_time, Carbon_monoxide_concentration,
label="Carbon monoxide concentration", linestyle="-")
# Show each point
plt.scatter(point_of_time, temperature, s=10)
plt.scatter(point_of_time, relative_humidity, s=10)
plt.scatter(point_of_time, Gas_concentration, s=10)
plt.scatter(point_of_time, Carbon_monoxide_concentration, s=10)
plt.rcParams['font.sans-serif'] = ['SimHei'] # Used to display Chinese labels normally
plt.rcParams['axes.unicode_minus'] = False # Used to display negative signs normally
plt.title("Coal mine sampling data sheet", fontsize=18)
plt.xlabel("Sampling time point", fontsize=12)
plt.ylabel("numerical value", fontsize=12)
plt.legend(fontsize=10)
plt.savefig('Line chart.jpg')
plt.show()
# Encapsulate all required functions in the main () method
def main(file):
# The type here is converted to string type, because float or int is not suitable for null and numeric values
data = np.loadtxt(file, skiprows=1, dtype=str, delimiter=',')
CoordinateArray = Create_Fill_Value_Arr(data) # Returns an array of coordinates containing all null values
for i in range(len(CoordinateArray)):
arr_final = Fill_Value(
data, int(CoordinateArray[i][0]), int(CoordinateArray[i][1])) # Although the floating-point type is converted to integer type, the coordinates here should be integers, so there is no problem with this forced conversion
arr_finally = CleaningData(arr_final)
CreatePicture(arr_finally)
return arr_finally
# It's best to use absolute addresses here, because some software seems to be inaccurate in locating relative addresses
file = r'task\Week 8\ug_detect.csv'
print(main(file))
Operation results
This is the modified result. The previous outliers and blank values have been re assigned
There is a small problem: the temperature has an abnormal value after calculation, which may remind me not to take it lightly. The code that feels super good will also be abnormal
[['1' '30.22' '69' '2.9' '3.6'] ['2' '37.68' '67.5' '2.86' '3.64'] ['3' '29.32' '66' '2.02' '1.66'] ['4' '37.44' '68' '1.18' '6.49'] ['5' '29.46' '75' '3.81' '4.78'] ['6' '30.12' '77' '1.93' '4.2'] ['7' '26.3' '83' '2.07' '3.67'] ['8' '30.340' '66' '1.46' '4.0299'] ['9' '34.38' '62' '3.36' '4.39'] ['10' '30.79' '75' '2.4' '5.79'] ['11' '25.17' '69' '3.34' '6.28'] ['12' '20.5' '80' '2.95' '6.22'] ['13' '37.21' '87.0' '21.569' '4.75'] ['14' '31.88' '94' '2.41' '1.49'] ['15' '39.94' '77' '1.98' '1.08'] ['16' '33.65' '67' '2.03' '1.79'] ['17' '27.21' '79' '1.41' '2.5'] ['18' '27.57' '107.0' '3.39' '1.7'] ['19' '31.59' '94' '3.27' '5.02'] ['20' '33.235' '93' '2.425' '4.69'] ['21' '34.88' '92' '1.58' '3.01'] ['22' '29.65' '92' '2.69' '5.16'] ['23' '26.05' '80' '2.61' '2.8'] ['24' '33.5' '79' '1.26' '6.81'] ['25' '34.71' '72' '1.82' '2.49'] ['26' '-41.04' '85.5' '3.77' '6.0900'] ['27' '37.06' '99' '1.94' '3.85'] ['28' '28.57' '64' '3.93' '3.125'] ['29' '28' '66' '1.08' '2.4'] ['30' '25.67' '60' '2.33' '3.84'] ['31' '20.59' '67' '3.88' '5.19'] ['32' '39.84' '107.0' '3.4' '3.77'] ['33' '29.22' '74' '1.09' '6.0900'] ['34' '33.19' '61' '3.22' '1.74'] ['35' '38.7' '62' '3.61' '6.52'] ['36' '30.130' '62' '3.62' '6.83'] ['37' '21.56' '93' '3.63' '4.43'] ['38' '38.06' '67' '3.21' '2.41'] ['39' '37.95' '68' '1.12' '4.56'] ['40' '35.8' '96' '1.52' '5.54'] ['41' '29.75' '80' '1.3' '2.23'] ['42' '32.69' '69' '3.26' '1.43'] ['43' '33.94' '96' '3.42' '1.4'] ['44' '36.31' '78' '3.69' '5.34']] [['1' '30.22' '69' '2.9' '3.6'] ['2' '37.68' '67.5' '2.86' '3.64'] ['3' '29.32' '66' '2.02' '1.66'] ['4' '37.44' '68' '1.18' '6.49'] ['5' '29.46' '75' '3.81' '4.78'] ['6' '30.12' '77' '1.93' '4.2'] ['7' '26.3' '83' '2.07' '3.67'] ['8' '30.340' '66' '1.46' '4.0299'] ['9' '34.38' '62' '3.36' '4.39'] ['10' '30.79' '75' '2.4' '5.79'] ['11' '25.17' '69' '3.34' '6.28'] ['12' '20.5' '80' '2.95' '6.22'] ['13' '37.21' '87.0' '21.569' '4.75'] ['14' '31.88' '94' '2.41' '1.49'] ['15' '39.94' '77' '1.98' '1.08'] ['16' '33.65' '67' '2.03' '1.79'] ['17' '27.21' '79' '1.41' '2.5'] ['18' '27.57' '107.0' '3.39' '1.7'] ['19' '31.59' '94' '3.27' '5.02'] ['20' '33.235' '93' '2.425' '4.69'] ['21' '34.88' '92' '1.58' '3.01'] ['22' '29.65' '92' '2.69' '5.16'] ['23' '26.05' '80' '2.61' '2.8'] ['24' '33.5' '79' '1.26' '6.81'] ['25' '34.71' '72' '1.82' '2.49'] ['26' '-41.04' '85.5' '3.77' '6.0900'] ['27' '37.06' '99' '1.94' '3.85'] ['28' '28.57' '64' '3.93' '3.125'] ['29' '28' '66' '1.08' '2.4'] ['30' '25.67' '60' '2.33' '3.84'] ['31' '20.59' '67' '3.88' '5.19'] ['32' '39.84' '107.0' '3.4' '3.77'] ['33' '29.22' '74' '1.09' '6.0900'] ['34' '33.19' '61' '3.22' '1.74'] ['35' '38.7' '62' '3.61' '6.52'] ['36' '30.130' '62' '3.62' '6.83'] ['37' '21.56' '93' '3.63' '4.43'] ['38' '38.06' '67' '3.21' '2.41'] ['39' '37.95' '68' '1.12' '4.56'] ['40' '35.8' '96' '1.52' '5.54'] ['41' '29.75' '80' '1.3' '2.23'] ['42' '32.69' '69' '3.26' '1.43'] ['43' '33.94' '96' '3.42' '1.4'] ['44' '36.31' '78' '3.69' '5.34']]
The box chart is to see whether there are abnormal values and how many abnormal values there are
But look at the output pictures, you can see that you have only learned a little fur
 The following is a combination of line chart and scatter chart
The following is a combination of line chart and scatter chart