Because different dimensions may affect the observation results of data
Solution: Map all data to the same scale
1. Maximum Normalization: Mapping all data to 0-1
Suitable for situations with clear boundaries, greatly influenced by Outlier
xscale=x−xminxmax−xminx_{\text {scale}}=\frac{x-x_{\min }}{x_{\max }-x_{\min }}xscale=xmax−xminx−xmin
2. Standardization of mean variance:
Normalize all data into data with a mean of 0 and a variance of 1
It can be applied to situations where there are no obvious boundaries of data and there may be extreme data values. It also applies to cases where data have obvious boundaries.
xscale=x−xmeansx_{s c a l e}=\frac{x-x_{m e a n}}{s}xscale=sx−xmean
Code example: (written in jupyter notebook)
1. Data normalization
import numpy as np
import matplotlib.pyplot as plt
Maximum Normalization
x = np.random.randint(0, 100, size=100) # 0 to 100 vectors, a total of 100
x
The result is:
array([29, 19, 31, 31, 9, 59, 14, 76, 69, 34, 30, 97, 13, 96, 0, 52,
86, 96, 40, 39, 98, 70, 76, 84, 82, 95, 52, 94, 84, 59, 89, 11, 29, 2,
99, 69, 44, 48, 48, 92, 54, 46, 1, 43, 16, 52, 51, 26, 83, 94, 50,74, 55, 30, 18, 44, 36, 62, 8, 56, 12, 44, 27, 26, 43, 88, 86, 58,12, 39, 62, 31, 77, 23, 78, 0, 58, 97, 58, 38, 83, 56, 70, 72, 84,54, 75, 2, 71, 1, 84, 77, 5, 23, 50, 3, 1, 58, 74, 47])
(x-np.min(x))/(np.max(x)-np.min(x)) # Maximum Normalization
The result is:
array([0.95918367, 0.59183673, 0.92857143, 0.85714286, 0.34693878, 0.97959184, 0.63265306, 0.05102041, 0.09183673, 0.47959184, 0.69387755, 0.30612245, 0.09183673, 0.85714286, 0.21428571, 0.23469388, 0.90816327, 0. , 0.73469388, 0.90816327, 0.36734694, 0.69387755, 0.2755102 , 0.45918367, 0.56122449, 0.21428571, 0.31632653, 0.85714286, 0.64285714, 0.26530612, 0.41836735, 0.90816327, 1. , 0.82653061, 0.16326531, 0.10204082, 0.6122449 , 0.87755102, 0.03061224, 0.2244898 , 0.48979592, 0.56122449, 0.33673469, 0.8877551 , 0.74489796, 0.01020408, 0.89795918, 0.89795918, 0.12244898, 0.41836735, 0.24489796, 0.18367347, 0.59183673, 0.51020408, 0.26530612, 0.59183673, 0.81632653, 0.07142857, 0.42857143, 0.8877551 , 0.04081633, 0.6122449 , 0.3877551 , 0.92857143, 0.70408163, 0.67346939, 0.20408163, 0.30612245, 0.87755102, 0.87755102, 0.60204082, 0.86734694, 0.19387755, 0.02040816, 0.39795918, 0.83673469, 0.02040816, 0.94897959, 0.23469388, 0.16326531, 0.26530612, 0.48979592, 0.20408163, 1. , 0.04081633, 0.42857143, 0.87755102, 0.20408163, 0.04081633, 0.21428571, 0.79591837, 0.17346939, 0.43877551, 0.21428571, 0.54081633, 0.83673469, 0.79591837, 0.17346939, 0.70408163, 0.79591837])
x = np.random.randint(0, 100, (50, 2)) # A matrix of 50*2 dimensions, with values ranging from 0 to 100
x[:10, :] # Take the top 10 elements
The result is:
array([[80, 95],
[ 1, 70],
[18, 70],
[31, 86],
[48, 57],
[84, 68],
[90, 29],
[44, 60],
[18, 39],
[42, 55]])
x = np.array(x, dtype=float) # Convert to floating point number
x[:10, :]
The result is:
array([[80., 95.],
[ 1., 70.],
[18., 70.],
[31., 86.],
[48., 57.],
[84., 68.],
[90., 29.],
[44., 60.],
[18., 39.],
[42., 55.]])
x[:, 0] = (x[:, 0] - np.min(x[:, 0]))/(np.max(x[:, 0]) - np.min(x[:,
0])) # For column 0 (the first feature), normalize x [:, 1]= (x [:, 1] - np. min (x [:, 1],
1]))/(np.max(x[:, 1]) - np.min(x[:, 1])) # Maximum normalization of column 1 (second feature)
x[:10, :]
The result is:
array([[0.84210526, 0.96938776],
[0.01052632, 0.71428571],
[0.18947368, 0.71428571],
[0.32631579, 0.87755102],
[0.50526316, 0.58163265],
[0.88421053, 0.69387755],
[0.94736842, 0.29591837],
[0.46315789, 0.6122449 ],
[0.18947368, 0.39795918],
[0.44210526, 0.56122449]])
plt.scatter(x[:, 0], x[:, 1])
plt.show()
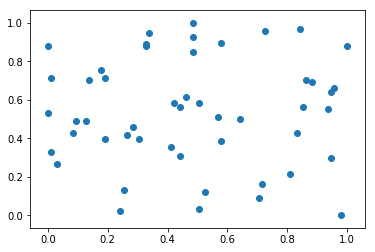
np.mean(x[:, 0]) # Means corresponding to column 0
The result is:
0.4785263157894737
np.std(x[:, 0]) # Variance corresponding to column 0
The result is:
0.3047560942923213
np.mean(x[:, 1]) # Means corresponding to column 1
The result is:
0.5375510204081633
np.std(x[:, 1]) # Variance corresponding to column 1
The result is:
0.2753547558441912
2. Standardization of Mean Variance Normalization
x2 = np.random.randint(0, 100, (50, 2)) # matrix
x2 = np.array(x2, dtype=float) # Converting to floating-point type
x2[:, 0] = (x2[:, 0] - np.mean(x2[:, 0]))/ np.std(x2[:, 0]) # Normalization of Mean Variance in Column 0
x2[:, 1] = (x2[:, 1] - np.mean(x2[:, 1]))/ np.std(x2[:, 1]) # Normalization of mean variance in column 1
plt.scatter(x2[:, 0], x[:, 1])
plt.show()
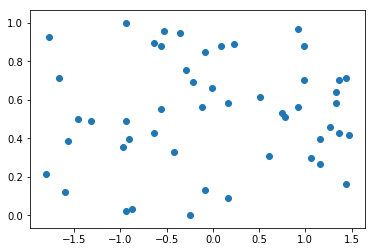
np.mean(x2[:, 0]) # Means corresponding to column 0
The result is:
-2.2204460492503132e-17
np.std(x2[:, 0]) # Variance corresponding to column 0
The result is:
0.9999999999999998
np.mean(x2[:, 1]) # Means corresponding to column 1
The result is:
-7.549516567451065e-17
np.std(x2[:, 1]) # Variance corresponding to column 1
The result is:
0.9999999999999999