In this paper, sklearn is used to predict the cumulative confirmed cases of new pneumonia. The main algorithms include linear regression, logical return, polynomial regression (quadratic curve, cubic curve, quartic curve, quintic curve), etc. specifically, prediction mainly includes the selection of algorithm. In many cases, the selection of algorithm is through the recall precision of data, training set, test set, and test The second is about the data set splitting, which needs to be divided into training set and test set for verification.
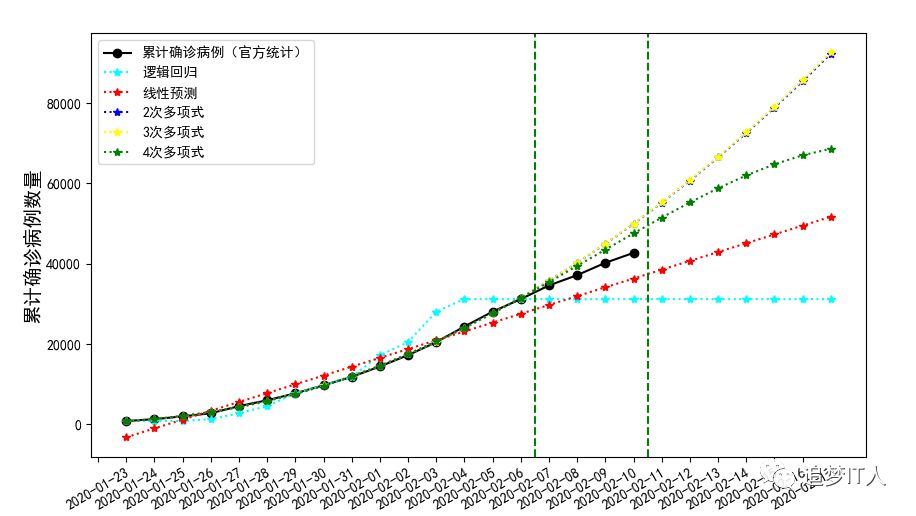
In general, there are only 15 records in the training set, 4 in the training set and 7 in the prediction. From the open chart, there are 2 fluctuations, so the effect of this model is average.
As follows:
import operator
import matplotlib.pyplot as plt
import numpy as np
from sklearn.preprocessing import PolynomialFeatures
from sklearn.linear_model import LinearRegression,LogisticRegression
import matplotlib.pyplot as plt
def init_data():
#Raw data
countrydatahistorys=[{'date': '2020-02-10', 'confirmedNum': 42708, 'suspectedNum': 21675, 'curesNum': 3998, 'deathsNum': 1017, 'suspectedIncr': 3536}, {'date': '2020-02-09', 'confirmedNum': 40224, 'suspectedNum': 23589, 'curesNum': 3283, 'deathsNum': 909, 'suspectedIncr': 4008}, {'date': '2020-02-08', 'confirmedNum': 37162, 'suspectedNum': 28942, 'curesNum': 2651, 'deathsNum': 812, 'suspectedIncr': 3916}, {'date': '2020-02-07', 'confirmedNum': 34594, 'suspectedNum': 27657, 'curesNum': 2052, 'deathsNum': 723, 'suspectedIncr': 4214}, {'date': '2020-02-06', 'confirmedNum': 31197, 'suspectedNum': 26359, 'curesNum': 1542, 'deathsNum': 637, 'suspectedIncr': 4833}, {'date': '2020-02-05', 'confirmedNum': 28060, 'suspectedNum': 24702, 'curesNum': 1153, 'deathsNum': 564, 'suspectedIncr': 5328}, {'date': '2020-02-04', 'confirmedNum': 24363, 'suspectedNum': 23260, 'curesNum': 892, 'deathsNum': 491, 'suspectedIncr': 3971}, {'date': '2020-02-03', 'confirmedNum': 20471, 'suspectedNum': 23214, 'curesNum': 630, 'deathsNum': 425, 'suspectedIncr': 5072}, {'date': '2020-02-02', 'confirmedNum': 17238, 'suspectedNum': 21558, 'curesNum': 475, 'deathsNum': 361, 'suspectedIncr': 5173}, {'date': '2020-02-01', 'confirmedNum': 14411, 'suspectedNum': 19544, 'curesNum': 328, 'deathsNum': 304, 'suspectedIncr': 4562}, {'date': '2020-01-31', 'confirmedNum': 11821, 'suspectedNum': 17988, 'curesNum': 243, 'deathsNum': 259, 'suspectedIncr': 5019}, {'date': '2020-01-30', 'confirmedNum': 9720, 'suspectedNum': 15238, 'curesNum': 171, 'deathsNum': 213, 'suspectedIncr': 4812}, {'date': '2020-01-29', 'confirmedNum': 7736, 'suspectedNum': 12167, 'curesNum': 124, 'deathsNum': 170, 'suspectedIncr': 4148}, {'date': '2020-01-28', 'confirmedNum': 5997, 'suspectedNum': 9239, 'curesNum': 103, 'deathsNum': 132, 'suspectedIncr': 3248}, {'date': '2020-01-27', 'confirmedNum': 4535, 'suspectedNum': 6973, 'curesNum': 51, 'deathsNum': 106, 'suspectedIncr': 2077}, {'date': '2020-01-26', 'confirmedNum': 2761, 'suspectedNum': 5794, 'curesNum': 49, 'deathsNum': 80, 'suspectedIncr': 3806}, {'date': '2020-01-25', 'confirmedNum': 1985, 'suspectedNum': 2684, 'curesNum': 38, 'deathsNum': 56, 'suspectedIncr': 1309}, {'date': '2020-01-24', 'confirmedNum': 1297, 'suspectedNum': 1965, 'curesNum': 38, 'deathsNum': 41, 'suspectedIncr': 1118}, {'date': '2020-01-23', 'confirmedNum': 830, 'suspectedNum': 1072, 'curesNum': 34, 'deathsNum': 25, 'suspectedIncr': 680}]
#Sort national trend data by time
countrydatahistorys=sorted(countrydatahistorys, key=operator.itemgetter('date'))
#Data needed to structure national provincial charts
xdata=list(range(len(countrydatahistorys)))
xlabel=list(row['date'] for row in countrydatahistorys)
#Add the independent variable X to forecast the next week
xdata.extend(list(range(19, 26)))
from datetime import date, datetime, timedelta
start_date = date(2020, 2, 11)
xlabel.extend(list(str(start_date + timedelta(i)) for i in range(7)))
#Generate cumulative diagnosis data, i.e. y value
confirmedNum=list(row['confirmedNum'] for row in countrydatahistorys)
suspectedNum=list(row['suspectedNum'] for row in countrydatahistorys)
#Transform data format to generate training set, test set and prediction set
Xlabel=np.array(xlabel).reshape(-1, 1)
X=np.array(xdata).reshape(-1, 1)
y=np.array(confirmedNum).reshape(-1, 1)
X_train=X[:15]
X_test=X[15:19]
X_predict=X[19:]
y_train=y[:15]
y_test=y[15:19]
return X_train,X_test,y_train,y_test,X_predict,X,y,xlabel
X_train,X_test,y_train,y_test,X_predict,X,y,Xlabel=init_data()
lr=LinearRegression().fit(X_train,y_train)
coef=lr.coef_
intercept=lr.intercept_
score_train=lr.score(X_train,y_train)
score_test=lr.score(X_test,y_test)
y_predict=lr.predict(X_test)
#--------------------------------
lg=LogisticRegression(C=0.2)
lg.fit(X_train,y_train)
y_lg_predict=lg.predict(X)
print('y_lg_predict=',y_lg_predict)
#--------------------------------
poly1 =PolynomialFeatures(degree=1)
X_ploy =poly1.fit_transform(X_train)
l1=LinearRegression()
l1.fit(X_ploy,y_train)
#--------------------------------
poly2 =PolynomialFeatures(degree=2)
X_ploy =poly2.fit_transform(X_train)
l2=LinearRegression()
l2.fit(X_ploy,y_train)
#--------------------------------
poly3 =PolynomialFeatures(degree=3)
X_ploy =poly3.fit_transform(X_train)
l3=LinearRegression()
l3.fit(X_ploy,y_train)
#--------------------------------
poly4 =PolynomialFeatures(degree=4)
X_ploy =poly4.fit_transform(X_train)
l4=LinearRegression()
l4.fit(X_ploy,y_train)
#--------------------------------
poly5 =PolynomialFeatures(degree=5)
X_ploy =poly5.fit_transform(X_train)
l5=LinearRegression()
l5.fit(X_ploy,y_train)
#--------------------------------
poly6 =PolynomialFeatures(degree=6)
X_ploy =poly6.fit_transform(X_train)
l6=LinearRegression()
l6.fit(X_ploy,y_train)
fig=plt.figure(figsize=(10,5.5))
plt.rcParams['font.sans-serif']=['SimHei']
#Draw the actual value. Note that X and y are not equal. X training set plus test set is equal to the actual y value
plt.plot(np.vstack((X_train,X_test)),y,color='black',marker='o',linestyle='-',label='Cumulative confirmed cases (official statistics)')
plt.plot(X,y_lg_predict,color='cyan',marker='*',linestyle=':',label='logistic regression')
plt.plot(X,intercept+X*coef,color='red',marker='*',linestyle=':',label='linear prediction')
plt.plot(X,l2.predict(poly2.fit_transform(X)),color='blue',marker='*',linestyle=':',label='2 Sub polynomial')
plt.plot(X,l3.predict(poly3.fit_transform(X)),color='yellow',marker='*',linestyle=':',label='3 Sub polynomial')
plt.plot(X,l4.predict(poly4.fit_transform(X)),color='green',marker='*',linestyle=':',label='4 Sub polynomial')
#Set x-axis label and its font size
plt.xlabel('date',fontsize=14)
#Set y-axis label and its font size
plt.ylabel('Cumulative number of confirmed cases',fontsize=14)
#Set X-axis sequence label value
plt.xticks(X-1,Xlabel,rotation=30,fontsize=10)
#Add training set, test set and prediction set to divide vertical line
plt.axvline(x=14.5,linestyle='--',c="green")
plt.axvline(x=18.5,linestyle='--',c="green")
#Add forecast result data label for test set
# for x,y in zip(X_test.tolist(), y_predict.tolist()):
# plt.text(x[0],y[0],'{:5.0f}'.format(y[0]), fontsize=8)
#Show legend
plt.legend()
plt.show()
Long press QR code to focus on "dream IT man"


