1, Theoretical basis
1. Node coverage model
0 / 1 coverage model is adopted in this paper. Please refer to for specific description here.
2. Symbiotic biological search algorithm (SOS)
SOS algorithm imitates biological behavior, and finally forms three evolutionary stages of "mutually beneficial symbiosis", "favorable symbiosis" and "parasitism", guiding individuals to evolve gradually. The key operations of SOS algorithm are as follows:
(1) Population initialization
Suppose the population number is N N N. The upper and lower limits of individual search range are respectively U U U and L L 50. Generate randomly according to the following formula N N N initial individuals X i X_i Xi: X i = L + rand ( 1 , D ) × ( U − L ) (1) X_i=L+\text{rand}(1,D)×(U-L)\tag{1} Xi=L+rand(1,D) × (U − L)(1) where, X i X_i Xi , represents the second in the population i i i( i = 1 , 2 , ⋯ , N i=1,2,\cdots,N i=1,2,..., N) biological individuals, rand \text{rand} rand representation [ 0 , 1 ] [0,1] Random number between [0,1], D D D represents the dimension of the optimized problem.
(2) Mutually beneficial symbiosis
For each individual in the population
X
i
X_i
Xi, randomly select one other individual
X
j
(
i
,
j
∈
{
1
,
2
,
⋯
,
N
}
,
j
≠
i
)
X_j(i, j\in\{1,2,\cdots,N\},j≠i)
Xj (i,j ∈ {1,2,..., N},j = i) and itself
Through mutually beneficial symbiosis, 2 new individuals are generated:
{
X
i
new
=
X
i
+
rand
(
0
,
1
)
×
(
X
best
−
MV
×
BF
1
)
X
j
new
=
X
j
+
rand
(
0
,
1
)
×
(
X
best
−
MV
×
BF
2
)
(2)
\begin{dcases}X_{i\text{new}}=X_i+\text{rand}(0,1)×(X_{\text{best}}-\text{MV}×\text{BF}_1)\\X_{j\text{new}}=X_j+\text{rand}(0,1)×(X_{\text{best}}-\text{MV}×\text{BF}_2)\end{dcases}\tag{2}
{Xinew=Xi+rand(0,1) × (Xbest−MV × BF1)Xjnew=Xj+rand(0,1) × (Xbest−MV × BF2) (2) where,
rand
(
0
,
1
)
\text{rand}(0,1)
rand(0,1) is
[
0
,
1
]
[0,1]
Random number between [0,1];
X
best
X_{\text{best}}
Xbest , is the optimal individual under the current number of iterations;
MV
=
(
X
i
+
X
j
)
/
2
\text{MV}=(X_i+X_j)/2
MV=(Xi + Xj) / 2 is "mutually beneficial vector", representing the relationship characteristics between organisms;
BF
1
\text{BF}_1
BF1,
BF
2
\text{BF}_2
BF2 ¢ is the benefit factor, and 1 or 2 is randomly selected.
A new individual is generated by formula (2)
X
i
new
X_{i_\text{new}}
Xinew # and
X
j
new
X_{j_\text{new}}
Xjnew, through and
X
i
X_i
Xi # and
X
j
X_j
Xj) compare the fitness values and retain them finally
X
i
new
X_{i_\text{new}}
Xinew , and
X
i
X_i
Xi,
X
j
new
X_{j_\text{new}}
Xjnew and
X
j
X_j
The better individual in Xj , is replaced
X
i
X_i
Xi # and
X
j
X_j
Xj.
(3) Partial benefit symbiosis
Individuals in the population
X
i
X_i
Xi , carry out the strategy of "partial benefit symbiosis" according to the following formula:
X
i
new
=
X
i
+
rand
(
−
1
,
1
)
×
(
X
best
−
X
j
)
(3)
X_{i\text{new}}=X_i+\text{rand}(-1,1)×(X_{\text{best}}-X_j)\tag{3}
Xinew=Xi+rand(−1,1) × (Xbest − Xj) (3) where,
X
j
X_j
Xj ^ is the population with
X
i
X_i
Xi) different individuals, i.e
i
,
j
∈
{
1
,
2
,
⋯
,
N
}
,
j
≠
i
i,j\in\{1,2,\cdots,N\},j≠i
i,j∈{1,2,⋯,N},j=i;
X
best
X_{\text{best}}
Xbest , is the optimal individual in the current population;
rand
(
−
1
,
1
)
\text{rand}(-1,1)
rand(− 1,1) is
[
−
1
,
1
]
[-1,1]
A random number between [− 1,1]. An individual newly generated by equation (3)
X
i
new
X_{i\text{new}}
Xinew decides whether to replace after comparing the fitness values
X
i
X_i
Xi.
It can be seen from the strategy of partial benefit symbiosis: similar to the "mutually beneficial symbiosis" stage, each individual randomly selects one other individual from the population to interact with itself; the difference is that the interaction between the two only makes
X
i
X_i
Xi # benefits for
X
j
X_j
Xj) its own development has neither interests nor harm.
(4) Parasitism
Generate one or more random numbers (no more than the number of gene loci) in the definition domain and modify the individual X i X_i Xi ^ corresponding loci to form new individuals Parasite_Vector \text{Parasite\_Vector} Parasite_vector (p_v for short), and randomly selected individuals X j ( i , j ∈ 1 , 2 , ⋅ ⋅ ⋅ , N , j ≠ i ) X_j(i, j∈ {1,2,· · ·, N}, j≠i) Xj (i,j ∈ 1,2, ⋅⋅⋅, N,j = I) (called "host") for fitness value comparison. If the fitness value of P_V is better, then "host" X j X_j Xj # will be replaced by P_V, otherwise X j X_j Xj , will be considered as immune individuals to be retained.
2, Simulation experiment and analysis
1. Function test and numerical analysis
SOS is compared with FPA, WOA, MFO, SCA and GWO. Taking F1, F2, F16, F17, F25 and F26 in literature [1] as examples, the population size is set to 30, the maximum number of iterations is set to 500, and each algorithm operates 30 times independently. The results are shown as follows:
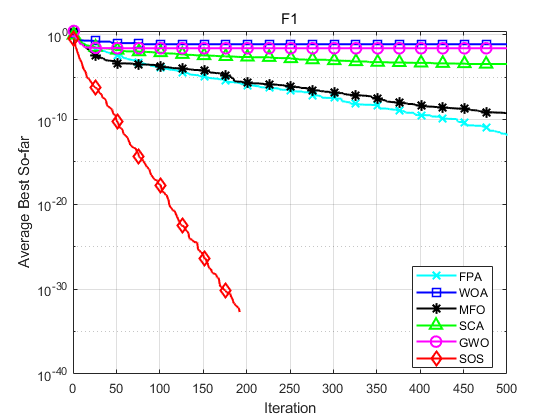
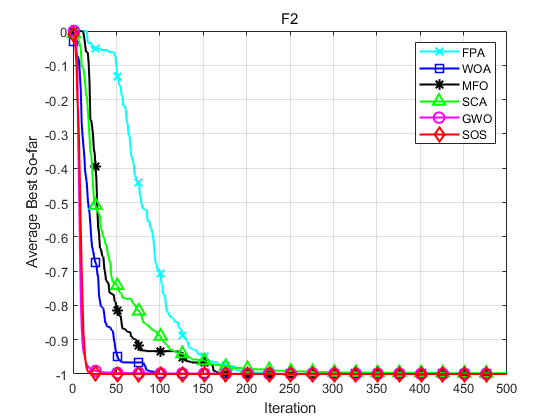
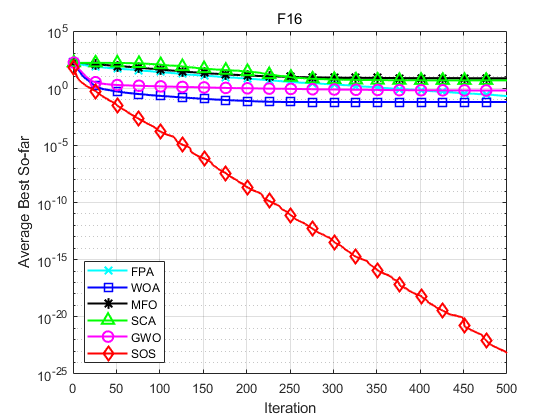
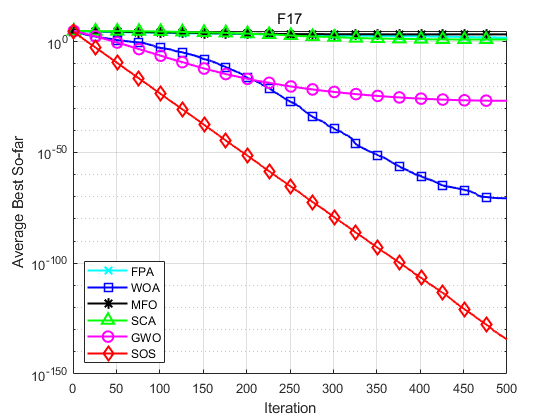


Function: F1 FPA: Worst value: 9.5109e-12,optimal value:1.1937e-18,average value:1.2781e-12,standard deviation:2.4481e-12 WOA: Worst value: 0.76207,optimal value:2.3163e-13,average value:0.076207,standard deviation:0.23253 MFO: Worst value: 1.6904e-08,optimal value:0,average value:5.6347e-10,standard deviation:3.0863e-09 SCA: Worst value: 0.0029332,optimal value:1.1219e-05,average value:0.00034985,standard deviation:0.0005416 GWO: Worst value: 0.76207,optimal value:3.8961e-09,average value:0.025402,standard deviation:0.13913 SOS: Worst value: 0,optimal value:0,average value:0,standard deviation:0 Function: F2 FPA: Worst value: -1,optimal value:-1,average value:-1,standard deviation:4.0834e-09 WOA: Worst value: -1,optimal value:-1,average value:-1,standard deviation:6.8516e-07 MFO: Worst value: -1,optimal value:-1,average value:-1,standard deviation:0 SCA: Worst value: -0.99207,optimal value:-0.99989,average value:-0.9979,standard deviation:0.0020357 GWO: Worst value: -1,optimal value:-1,average value:-1,standard deviation:8.2047e-07 SOS: Worst value: -1,optimal value:-1,average value:-1,standard deviation:0 Function: F16 FPA: Worst value: 0.4289,optimal value:0.087323,average value:0.20346,standard deviation:0.075546 WOA: Worst value: 0.15936,optimal value:0.024134,average value:0.061615,standard deviation:0.037101 MFO: Worst value: 42.6906,optimal value:0.001229,average value:7.4352,standard deviation:13.1045 SCA: Worst value: 6.976,optimal value:3.7828,average value:4.8192,standard deviation:0.61568 GWO: Worst value: 1.2637,optimal value:6.5089e-05,average value:0.62033,standard deviation:0.30985 SOS: Worst value: 5.5871e-23,optimal value:1.1133e-26,average value:6.7267e-24,standard deviation:1.3362e-23 Function: F17 FPA: Worst value: 130.5562,optimal value:37.1901,average value:68.9236,standard deviation:24.3054 WOA: Worst value: 5.9377e-70,optimal value:2.1857e-89,average value:1.9822e-71,standard deviation:1.084e-70 MFO: Worst value: 10001.9982,optimal value:0.66141,average value:2006.8331,standard deviation:4065.0722 SCA: Worst value: 68.3455,optimal value:0.0017531,average value:10.2897,standard deviation:17.2873 GWO: Worst value: 1.278e-26,optimal value:3.4061e-29,average value:2.0882e-27,standard deviation:3.1648e-27 SOS: Worst value: 1.0245e-133,optimal value:5.2837e-139,average value:4.0699e-135,standard deviation:1.8616e-134 Function: F25 FPA: Worst value: 2.3744,optimal value:1.3128,average value:1.6881,standard deviation:0.24307 WOA: Worst value: 0,optimal value:0,average value:0,standard deviation:0 MFO: Worst value: 90.8951,optimal value:0.3891,average value:16.0292,standard deviation:33.9355 SCA: Worst value: 1.473,optimal value:0.17989,average value:0.91643,standard deviation:0.34017 GWO: Worst value: 0.042361,optimal value:0,average value:0.0051044,standard deviation:0.009838 SOS: Worst value: 0,optimal value:0,average value:0,standard deviation:0 Function: F26 FPA: Worst value: 16.5721,optimal value:6.0727,average value:10.7366,standard deviation:3.0996 WOA: Worst value: 7.9936e-15,optimal value:8.8818e-16,average value:4.5593e-15,standard deviation:1.9755e-15 MFO: Worst value: 19.9631,optimal value:1.9986,average value:15.8598,standard deviation:5.9102 SCA: Worst value: 20.3871,optimal value:0.022724,average value:12.4272,standard deviation:9.5256 GWO: Worst value: 1.4655e-13,optimal value:7.5495e-14,average value:1.0617e-13,standard deviation:2.2479e-14 SOS: Worst value: 4.4409e-15,optimal value:8.8818e-16,average value:3.8488e-15,standard deviation:1.3467e-15
Experimental results show that SOS algorithm has better accuracy and obvious advantage of convergence speed.
2. SOS optimized WSN coverage
Set the monitoring area as
50
m
×
50
m
50 m×50 m
50m × 50m two-dimensional plane, number of sensor nodes
N
=
35
N=35
N=35, its sensing radius is
R
s
=
5
m
R_s=5m
Rs = 5m, communication radius
R
c
=
10
m
R_c=10m
Rc = 10m, 500 iterations. The evolution curves of initial deployment, SOS optimization coverage and SOS algorithm coverage are shown in the figure below.


 The node locations and corresponding coverage of initial deployment and final deployment are respectively:
The node locations and corresponding coverage of initial deployment and final deployment are respectively:
Initial position: 32.6545 32.446 41.2426 45.945 13.0846 49.5728 29.2021 43.8733 48.0295 3.955 47.3663 37.7734 1.3201 16.5851 19.2888 18.9047 12.7334 26.0067 22.0335 31.4901 23.0718 7.1101 47.1069 20.9824 37.1141 18.8479 9.1479 36.7856 14.2348 8.1498 13.6283 15.9827 41.5419 35.7073 40.6812 44.231 26.2897 40.0419 4.9575 40.1513 34.5246 27.7789 29.3381 33.8751 32.179 6.9931 9.0222 42.9654 17.4824 23.1989 4.3904 33.918 14.0562 35.1384 49.8881 42.8776 22.4286 23.2057 18.4397 44.4531 39.7413 45.4298 19 34.206 44.524 32.9813 6.6118 27.7741 43.4236 16.3376 Initial coverage: 0.69512 Optimal location: 15.7466 29.0815 10.9187 41.3071 47.0841 19.2871 35.9243 39.494 36.3785 25.5118 46.3233 4.6537 28.624 12.7572 35.1543 9.1547 20.18 19.4115 4.5097 2.912 4.2651 46.435 46.7193 27.6348 26.4977 4.1642 18.8016 45.9033 38.8681 32.5106 9.3462 31.4209 3.2417 36.6712 25.3966 48.3782 34.4429 47.0442 43.9273 13.483 45.667 45.6503 18.5576 36.9728 21.9133 28.5272 3.554 23.739 23.2557 13.3457 17.6117 1.143 37.6181 3.3859 29.5581 22.2684 10.7122 20.5166 16.3281 13.6298 37.6176 17.8293 6.3979 11.3552 46.7737 36.7952 29.6526 32.3906 26.8222 40.7059 Optimal coverage: 0.89773
The experimental results show that SOS can effectively improve the coverage of sensor nodes and make the node distribution more uniform.
3, References
[1] Min-Yuan Cheng, Doddy Prayogo. Symbiotic Organisms Search: A new metaheuristic optimization algorithm[J]. Computers and Structures, 2014, 139(jul.): 98-112.
[2] Wang Yanjiao, Ma Zhuang Elite symbiotic biological search algorithm based on sub population stretching operation . control and decision making, 2019, 34 (7): 1355-1364