Copyright notice: This is the original article of the blogger, following CC 4.0 BY-SA copyright agreement. Please attach the original source link and this notice for reprint.
This blog implements the https://disc.gsfc.nasa.gov/ The ozone, formaldehyde and nitrogen dioxide data downloaded from the website are processed with level 3 data. In this case, ozone data is processed. The processing flow of formaldehyde and nitrogen dioxide data is the same as that of ozone data.
1, Image download
PS: you must register the website before downloading and modifying data.
1. Follow the steps below and click the search button to search.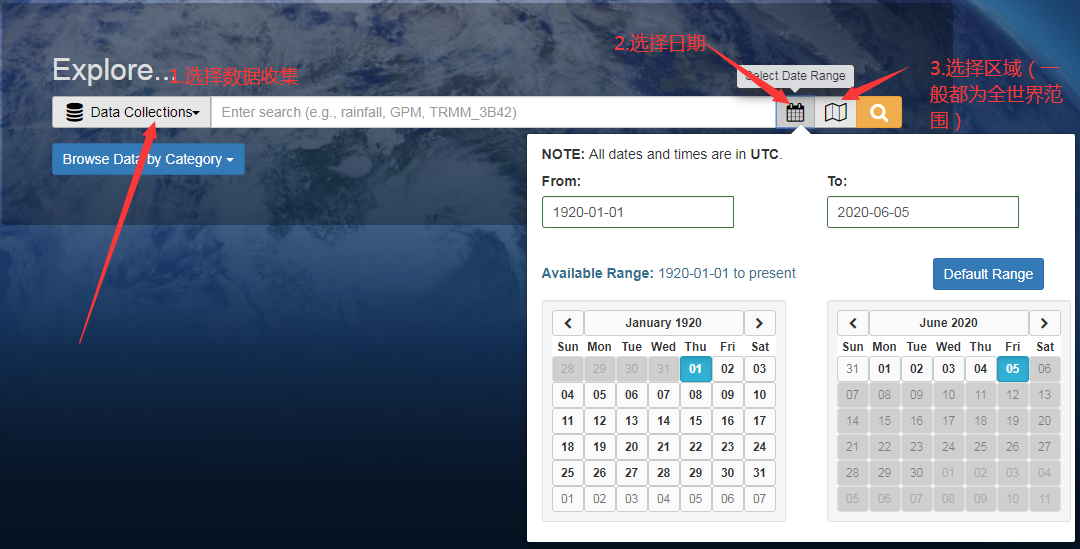
2. Search next
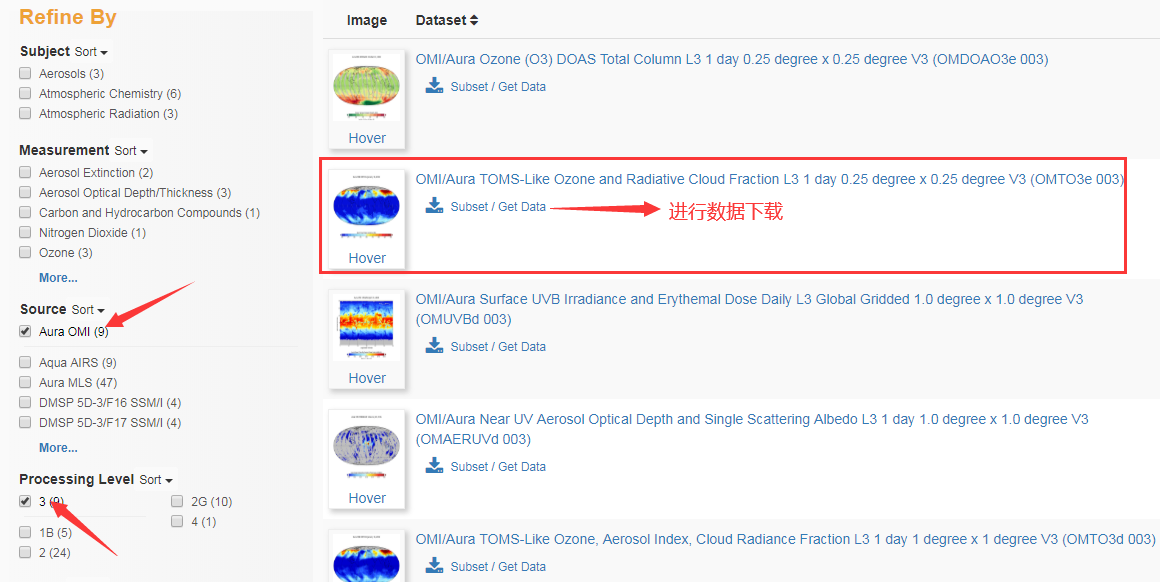
3. Take ozone data as an example. After clicking Subset/Get Data, because the date and area have been selected in the first step, click GetData directly,
4. Download the data. You can have a look at the user manual you are interested in.
Note: because it is daily average data, if you want to download in batch, please refer to the following link: https://www.cnblogs.com/huapiao-chen/articles/8901602.html
No personal test, because I have installed other versions of Firefox browser on this machine before, without using the link in the given network disk. If there is VPN when downloading, it is better to use VPN for downloading. If there is no VPN, the amount of ozone and nitrogen dioxide data is small, and the download speed can be seen in the past, but the amount of formaldehyde data is large (more than 60 MB). Wait slowly...
2, Data processing
Because it is L3 level data, it can be used only by using GLT file for correction.
Use IDL to preprocess the data
1. Read HDF file
file='E:\Ozone\OMI-Aura_L3-OMTO3e_2019m0706_v003-2020m0117t131443.he5' raster=e.OpenRaster(file,DATASET_NAME='/HDFEOS/GRIDS/OMI Column Amount O3/Data Fields/ColumnAmountO3');Read ozone data set
2. For its longitude and latitude information, we can get the storage mode of its longitude and latitude from its description document. The number of rows and columns of its longitude and latitude matrix is 720 * 1440, and the initial latitude value of the data is - 90 °, the initial longitude value is - 180 °, and the interval is 0.25 °. In this way, we can convert the longitude and latitude into the matrix with the same O3 concentration
data=raster.GetData(band=0)
dim=size(data,/dimension);Get data dimension information
rows=dim[1]
cols=dim[0]
lat=make_array(cols,rows);Same as O3 Latitude matrix of the same matrix
lon=make_array(cols,rows);Same as O3 Longitude matrix of the same matrix
for i=0,rows-1 do begin
for j=0,cols-1 do begin
lat[j,i]=-90+0.25*i
lon[j,i]=-180+0.25*j
endfor
endfor ;Equivalent to filling in the value of the matrix
lat_raster=ENVIRASTER(lat,URI='E:\Ozone\lat.dat')
lat_raster.save
lon_raster=ENVIRASTER(lon,uri='E:\Ozone\lon.dat')
lon_raster.save ;Output and save longitude and latitude data3. Make GLT file and correct the original data with GLT file. You can also use ENVI software to make GLT files. You can refer to the following links
http://blog.sina.com.cn/s/blog_764b1e9d0100qzow.html
;Go ahead GLT Production of documents.
latitude=e.openraster('E:\Ozone\lat.dat')
longitude=e.openraster('E:\Ozone\lon.dat');Read the longitude and latitude file just made
out_name='E:\Ozone\GLT.dat' ;Output path
lat_id=ENVIRASTERTOFID(latitude)
lon_id=ENVIRASTERTOFID(longitude) ;Store as identity
envi_file_query,lat_id,nb=nb_lat
y_pos=lindgen(nb_lat)
envi_file_query,lon_id,nb=nb_lon ;Query corresponding information
x_pos=lindgen(nb_lon)
input_prj=ENVI_PROJ_CREATE(/geographic,datum='WGS-84')
output_prj=ENVI_PROJ_CREATE(/geographic,datum='WGS-84');Set projection information
ENVI_DOIT,'ENVI_GLT_DOIT',dims=dims,I_PROJ=input_prj,O_PROJ=output_prj,$
OUT_NAME=out_name,ROTATION=0,X_FID=lon_id,$
X_POS=x_pos[0], Y_FID=lat_id, Y_POS=y_pos[0] ;conduct GLT Production of
;Based on GLT Correction of documents
GLT_raster=e.openraster('E:\Ozone\GLT.dat')
glt_fid=envirastertofid(GLT_RASTER)
raster=e.openraster(file,DATASET_NAME='/HDFEOS/GRIDS/OMI Column Amount O3/Data Fields/ColumnAmountO3') ;Open dataset
out_path=''E:\Ozone\20190706.dat'
raster_id=envirastertofid(raster) ;Store as identity
envi_file_query,raster_id,ns=ns,nb=nb
pos=indgen(nb) ;Get the information you need
envi_doit,'ENVI_GEOREF_FROM_GLT_DOIT',FID=raster_id,GLT_DIMS=dims,GLT_FID=glt_fid,out_name=out_path,pos=pos ;Make correctionsThe results are as follows:
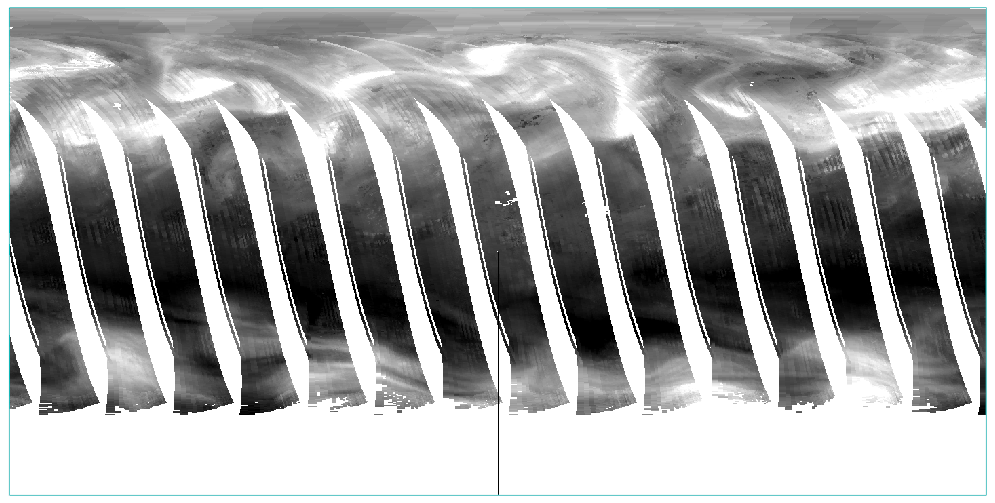
4. Achievement display
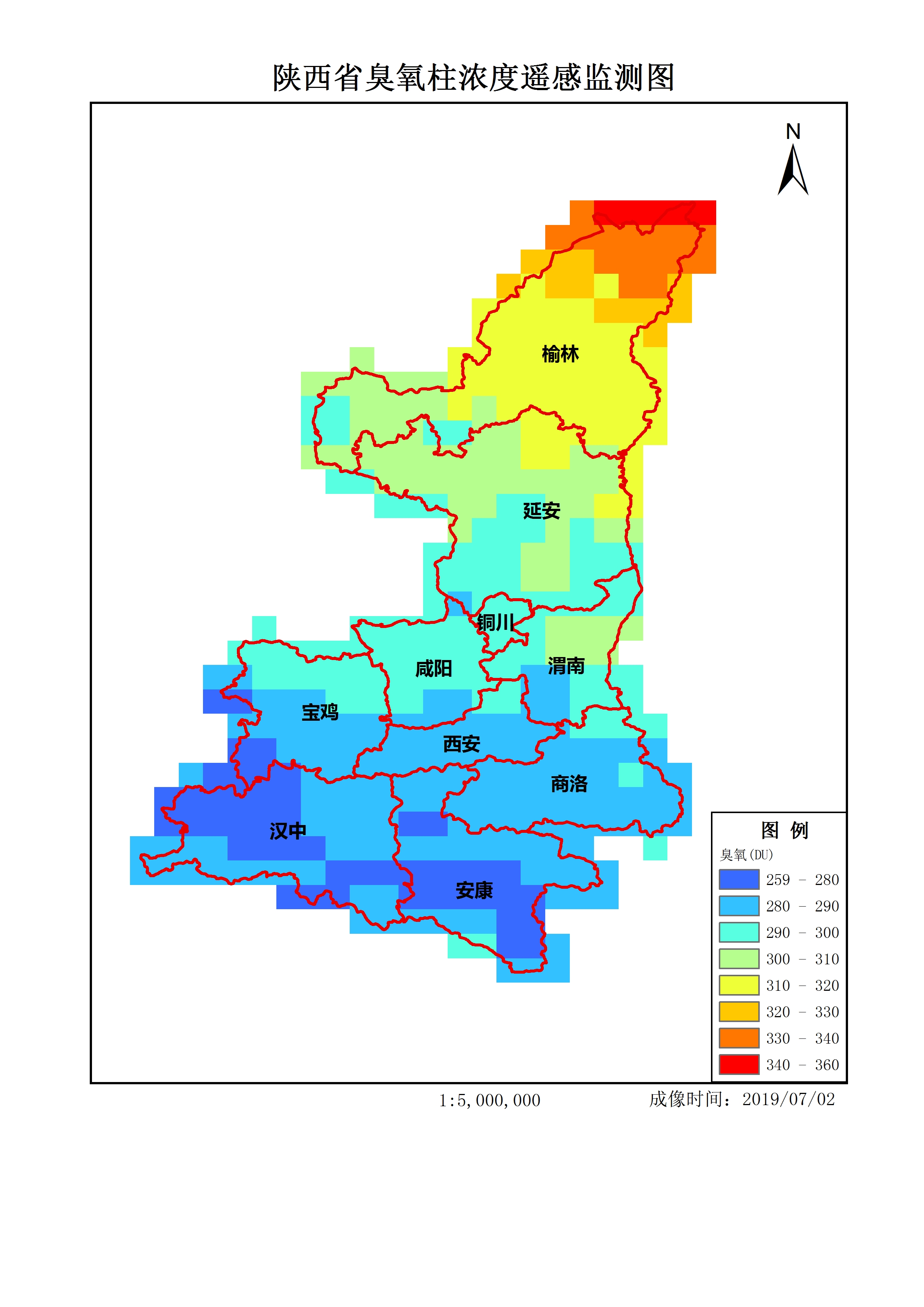
Make GIF.
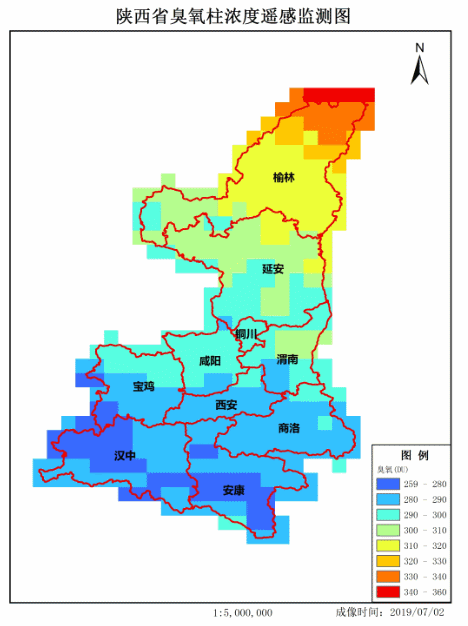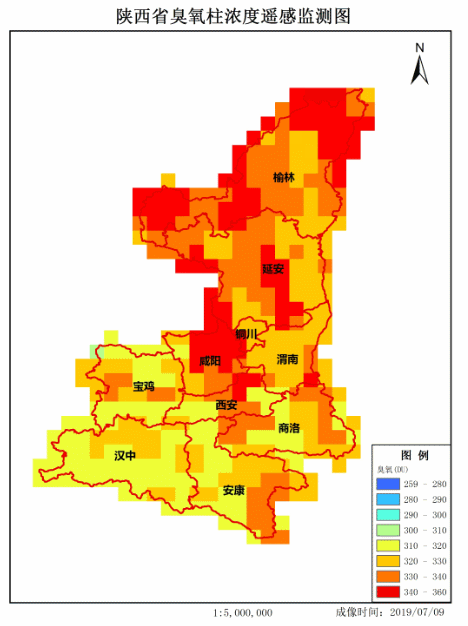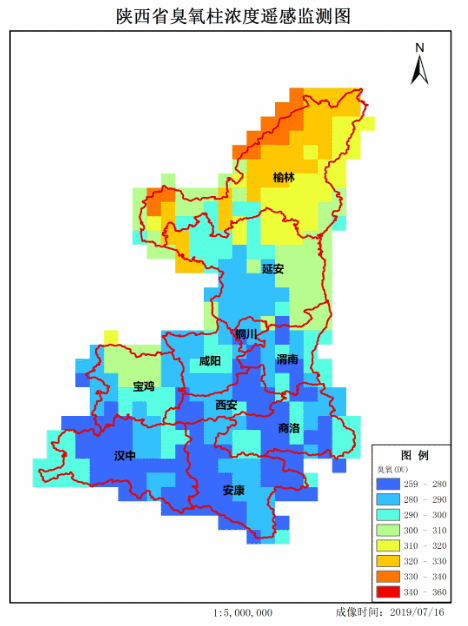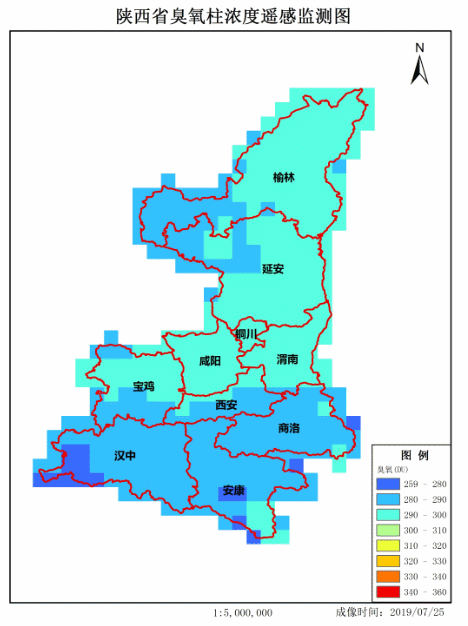
If there are any mistakes, please correct them. Thank you very much!
PS: with batch code.