Article directory
problem
solution
As a little white, I really did not find any suitable way, so I can only consult the papers of the big guys, found a very good one, and retailed it.
The original link is as follows
http://www.yndxxb.ynu.edu.cn/yndxxb/article/2018/0258-7971-40-5-879.html#outline_anchor_7
1 Data analysis
1.1 Data Visualization
This part is not very difficult, that is, reading data and drawing.
1.1.1 Read all data and draw pictures
To read all the data, just open excel and take out the contents of the corresponding fields. We need to pay attention to the processing of time type.
data = xlrd.open_workbook("2018 Northeast Three Provinces Mathematical Modeling League F Annex.xls") # Open excel
table = data.sheet_by_name("Lambert 20140 415-0612") # Read sheet
times = []
flow1 = []
flow2 = []
for i in range(1, table.nrows):
row = table.row_values(i) # Line data in an array
date = xldate_as_tuple(row[0], 0) # Date tuple
times.append(datetime.datetime(*date))
flow1.append(row[1])
flow2.append(row[2])
draw_all(times, flow1, flow2)
Drawing directly from the read data still requires attention to the processing of time types.
def draw_all(times, flow1, flow2):
# Configure abscissa
length = len(times)
coordinate = [times[0], times[int(length / 4)], times[int(2 * length / 4)], times[int(3 * length / 4)],
times[length - 1]]
plt.xticks(coordinate)
plt.gca().xaxis.set_major_formatter(mdates.DateFormatter('%m-%d'))
# Configure title, axis name
plt.title("ALL DAY")
plt.xlabel("time")
plt.ylabel("flow (L/s)")
# Plot
plt.plot(times, flow1, label='DMA1')
plt.plot(times, flow2, label='DMA2')
plt.legend(loc=4) # Specify the location of legend so that readers can help themselves with its usage
plt.gcf().autofmt_xdate() # Automatic Rotation Date Marker
plt.show()
give the result as follows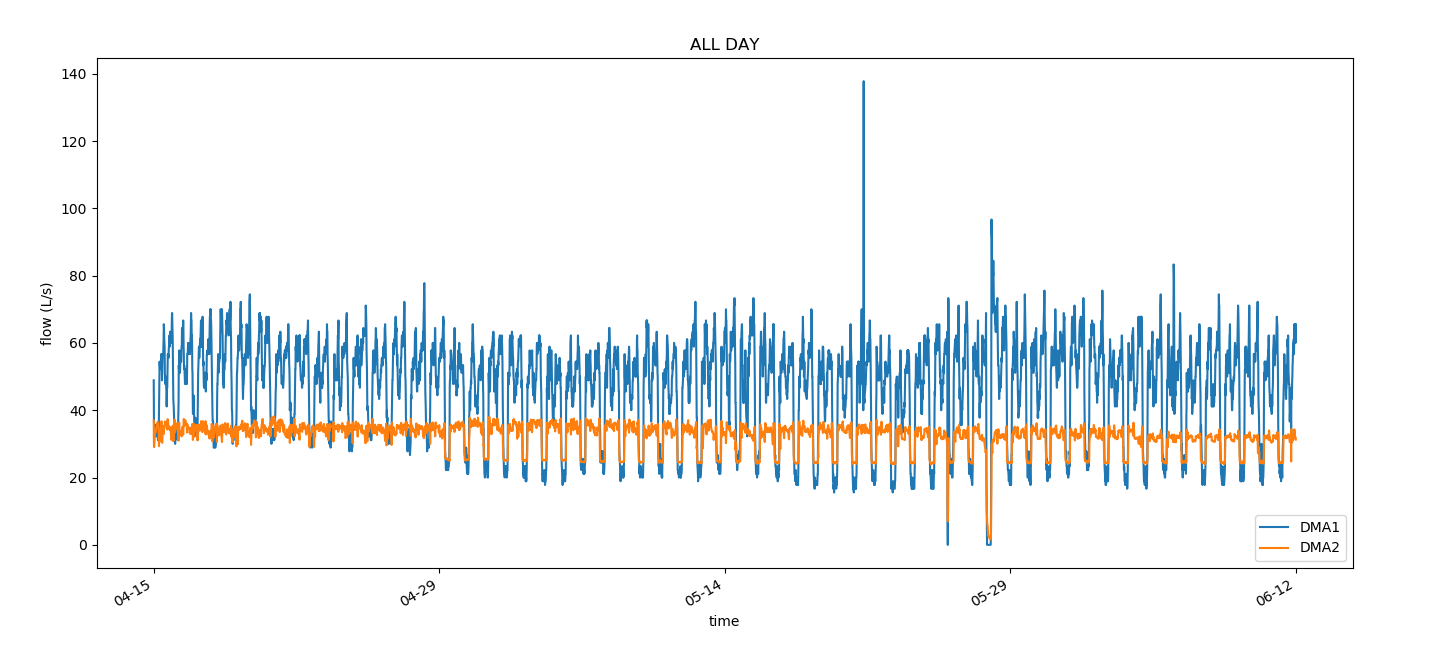
1.1.2 Read the data of a certain day and draw a picture
Notice here how to determine the date of reading the data as specified.
def read_oneday(month, day):
'''
:param month: Designated month
:param day: Designated days
:return: There is no return value, but the drawing method is then invoked. May also times,flow Return to the main function call
'''
data = xlrd.open_workbook("2018 Northeast Three Provinces Mathematical Modeling League in 1997 F Annex.xls") # Open excel
table = data.sheet_by_name("Lambert 20140 415-0612") # Read sheet
times = []
flow1 = []
flow2 = []
for i in range(1, table.nrows):
row = table.row_values(i) # Line data in an array
date = xldate_as_tuple(row[0], 0)
if date[1] == month and date[2] == day:
times.append(datetime.datetime(*date))
flow1.append(row[1])
flow2.append(row[2])
draw_oneday(times, flow1, flow2)
Compared with the previous drawing, the coordinate axes and headings need to be modified, and attention should still be paid to date processing.
def draw_oneday(times, flow1, flow2):
# Configuration abscissa
length = len(times)
coordinate = [times[0], times[int(length / 4)], times[int(2 * length / 4)], times[int(3 * length / 4)],
times[length - 1]]
plt.xticks(coordinate)
plt.gca().xaxis.set_major_formatter(mdates.DateFormatter('%H:%M:%S'))
# Configure title, axis name
plt.title(times[0].strftime('%Y/%m/%d'))
plt.xlabel("time")
plt.ylabel("flow (L/s)")
# Plot
plt.plot(times, flow1, label='DMA1')
plt.plot(times, flow2, label='DMA2')
plt.legend(loc=4) # Specify the location of legend so that readers can help themselves with its usage
plt.gcf().autofmt_xdate() # Automatic Rotation Date Marker
plt.show()
give the result as follows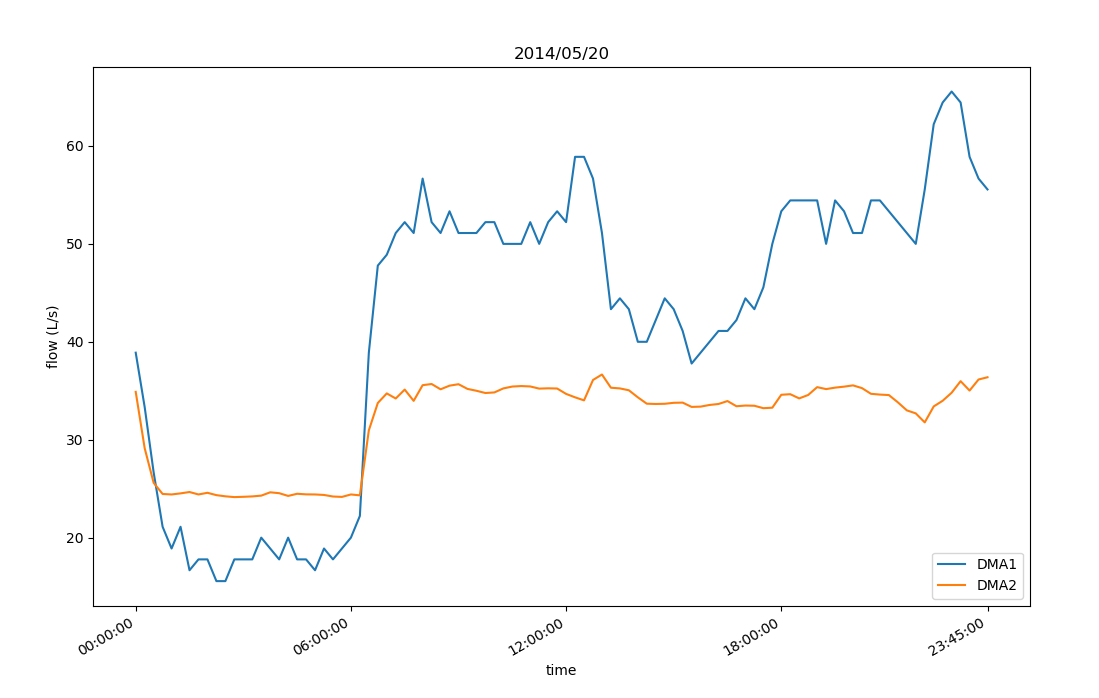
1.2 Water Flow
Find out the daily water leakage first.
def find_leak():
data = xlrd.open_workbook("2018 Northeast Three Provinces Mathematical Modeling League in 1997 F Annex.xls") # Open excel
table = data.sheet_by_name("Lambert 20140 415-0612") # Read sheet
# Leakage, dictionary type, key date, value for the day of leakage
leak1 = {}
leak2 = {}
# Minimum flow at 2-5 points of the day as leakage
minFlow1 = 999
minFlow2 = 999
for i in range(1, table.nrows):
row = table.row_values(i) # Line data in an array
date = xldate_as_tuple(row[0], 0)
if i != 1 and date[2] != oldDate[2]: # Compare date changes
key = str(oldDate[1])+"-"+str(oldDate[2])
leak1[key] = minFlow1
leak2[key] = minFlow2
minFlow1 = 999
minFlow2 = 999
if date[3] >= 2 and date[3] <= 5:
minFlow1 = min(minFlow1, row[1])
minFlow2 = min(minFlow2, row[2])
oldDate = date # Record a date
if i == table.nrows - 1: ##Deal with the last day alone
key = str(date[1]) + "-" + str(date[2])
leak1[key] = minFlow1
leak2[key] = minFlow2
return leak1, leak2
Calculate water consumption according to water leakage, pay attention to replacing negative number with 0.
def cal_usewater():
data = xlrd.open_workbook("result.xls") # Open excel
table = data.sheet_by_name("Lambert 20140 415-0612") # Read sheet
from xlutils.copy import copy
wb = xlutils.copy.copy(data)
sheet = wb.get_sheet(0)
sheet.write(0, 3, 'use1')
sheet.write(0, 4, 'use2')
leak1, leak2 = find_leak()
for i in range(1, table.nrows):
row = table.row_values(i) # Line data in an array
date = xldate_as_tuple(row[0], 0)
key = str(date[1]) + "-" + str(date[2])
sheet.write(i, 3, str(max(row[1] - leak1[key], 0)))
sheet.write(i, 4, str(max(row[2] - leak2[key], 0)))
wb.save("result.xls")
Because the result is too long, only part is shown. Remember to copy the original document and append the result directly to the file for easy reuse.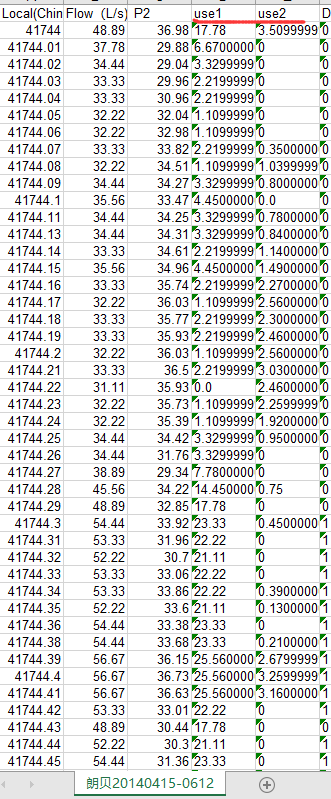
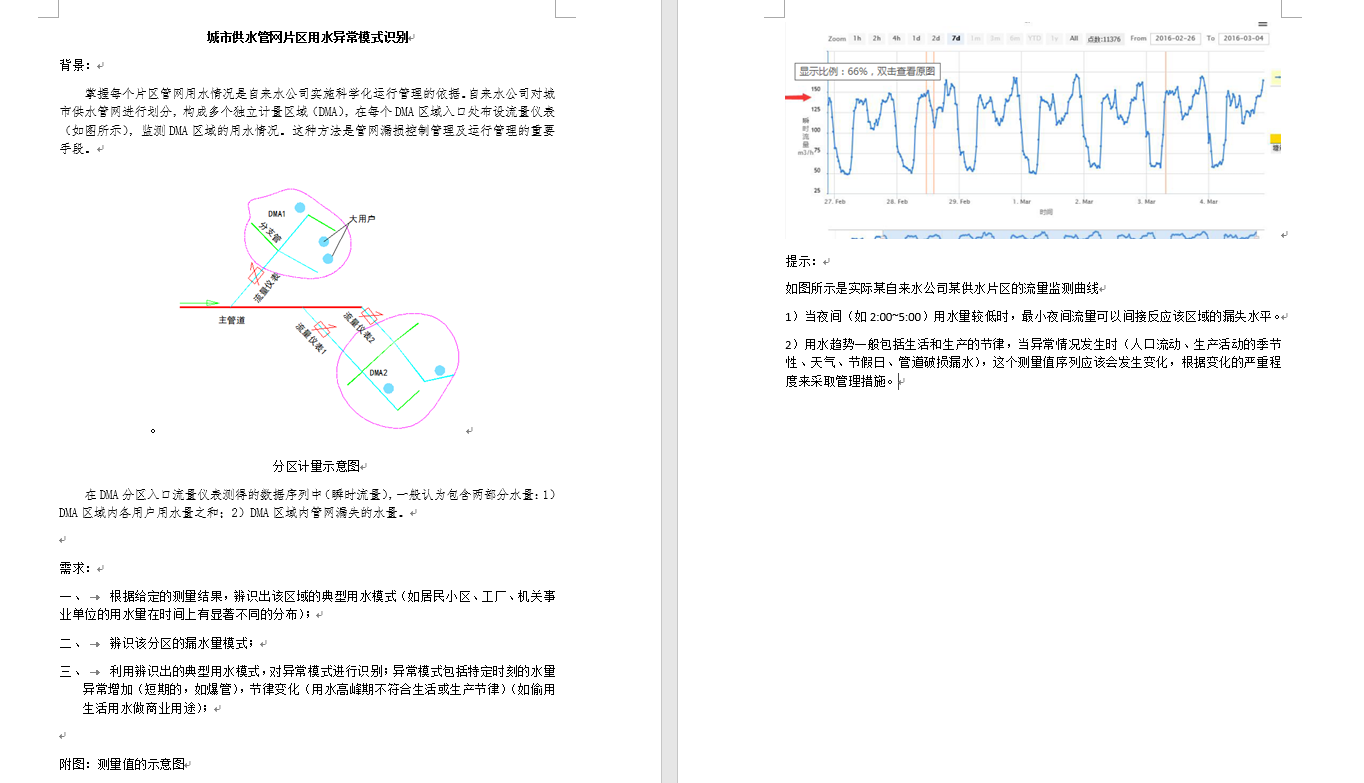
2 Typical Water Use Model
2.1 Unithermal Coding
This step maps the sample set to a Euclidean space for easy distance calculation, where the time characteristics are unithermal coded with constant water flow.
2.1.1 Mapping Date
def get_timeMap():
data = xlrd.open_workbook("2018 Northeast Three Provinces Mathematical Modeling League in 1997 F Annex.xls") # Open excel
table = data.sheet_by_name("Lambert 20140 415-0612") # Read sheet
timeMap = {}
for i in range(1, table.nrows):
row = table.row_values(i) # Line data in an array
date = xldate_as_tuple(row[0], 0)
if date[1] == 4 and date[2] == 15:
key = str(date[3]) + ":" + str(date[4]) + ":" +str(date[5])
timeMap[key] = i
enc = preprocessing.OneHotEncoder()
studyData = []
for item in timeMap.values():
studyData.append([item])
enc.fit(studyData) # fit to learn coding
endcodeData = enc.transform(studyData).toarray().tolist() # Coding
count = 0
for key in timeMap.keys():
timeMap[key] = endcodeData[count]
count = count + 1
return timeMap
2.1.2 Coding Vector
A sample vector is constructed by combining the mapping date and water consumption.
timeMap = get_timeMap()
data = xlrd.open_workbook("2018 Northeast Three Provinces Mathematical Modeling League F Annex.xls") # Open excel
table = data.sheet_by_name("Lambert 20140 415-0612") # Read sheet
vectors1 = []
vectors2 = []
for i in range(1, table.nrows):
row = table.row_values(i) # Line data in an array
date = xldate_as_tuple(row[0], 0)
key = str(date[3]) + ":" + str(date[4]) + ":" + str(date[5])
item1 = copy.deepcopy(timeMap[key]) # Pay attention to deep replication
item1.append(use1[i-1])
vectors1.append(item1)
item2 = copy.deepcopy(timeMap[key])
item2.append(use2[i - 1])
vectors2.append(item2)
return np.array(vectors1), np.array(vectors2)
give the result as follows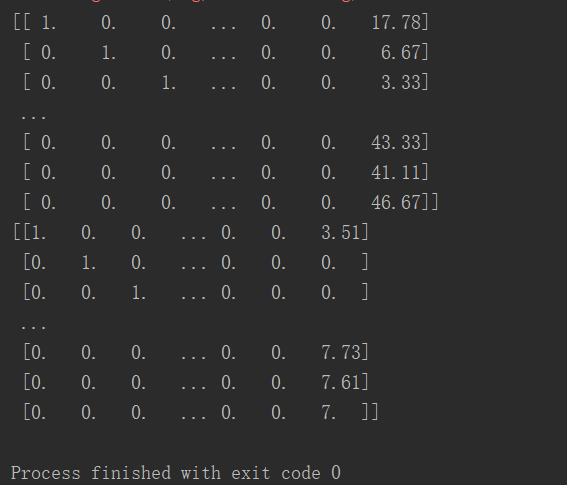
2.2 Cluster Evaluation
Take contour coefficient
def choose_by_silhouette(X):
tests = [2, 3, 4, 5]
sc_scores = []
for k in tests:
kmeans_model = KMeans(n_clusters=k).fit(X)
sc_score = metrics.silhouette_score(X, kmeans_model.labels_, metric='euclidean')
sc_scores.append(sc_score)
return sc_scores
The results of k taking 2, 3, 4 and 5 are as follows.
2.3 K Mean Classification
You need to retrieve the list of times before classifying, using the previous read_all() method, but you need to change the date in times to the same day.
for count in range(0, len(times)):
times[count] = times[count].replace(2014, 4, 15)
The parameters obtained before input can be classified.
def cluster_by_K(x1, x2, X, k, name):
'''
:param x1: Time list
:param x2: Water consumption list
:param X: Matrix composed of sample vectors
:param k: k value
:param name: Picture name
:return:
'''
data = xlrd.open_workbook("result.xls") # Open excel
from xlutils.copy import copy
wb = xlutils.copy.copy(data)
sheet = wb.get_sheet(0)
if name == 'DMA1':
cloumn = 3
else:
cloumn = 4
sheet.write(0, cloumn, name)
plt.figure(figsize=(8, 6))
colors = ['b', 'g', 'r', 'c', 'm', 'y', 'k', 'b']
markers = ['o', 's', 'D', 'v', '^', 'p', '*', '+']
kmeans_model = KMeans(n_clusters=k).fit(X)
for i, l in enumerate(kmeans_model.labels_):
plt.subplot(k, 1, 1 + l)
sheet.write(i+1, cloumn, str(l))
print(i)
plt.plot(x1[i], x2[i], color=colors[l], marker=markers[l])
wb.save("result.xls")
for t in range(1, k+1):
plt.subplot(k, 1, t)
plt.gca().xaxis.set_major_formatter(mdates.DateFormatter('%H:%M:%S'))
plt.gcf().autofmt_xdate() # Automatic Rotation Date Marker
plt.title('The first'+str(t)+'species', fontproperties=font)
plt.savefig(name+'.png')
return kmeans_model.labels_
The results of DMA1 are as follows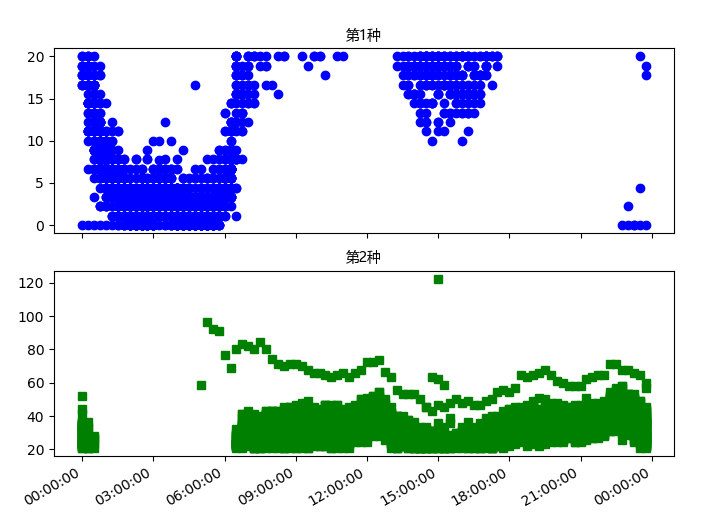
The results of DMA2 are as follows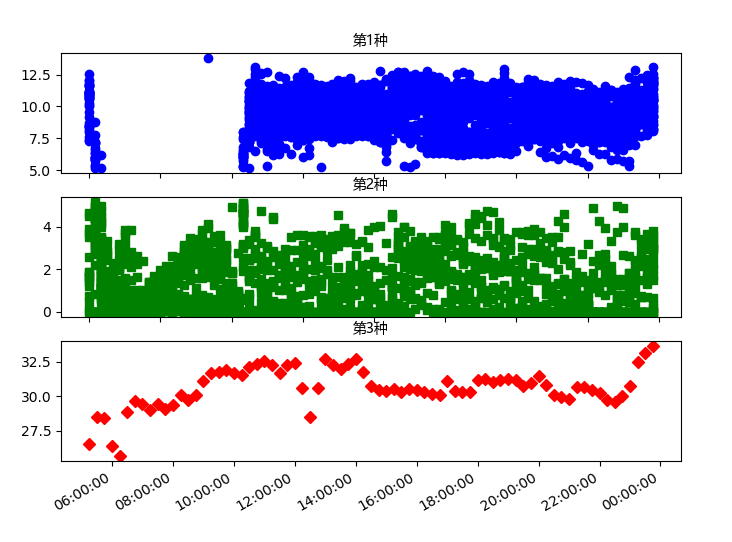
At the same time, the results are appended to the document for reuse.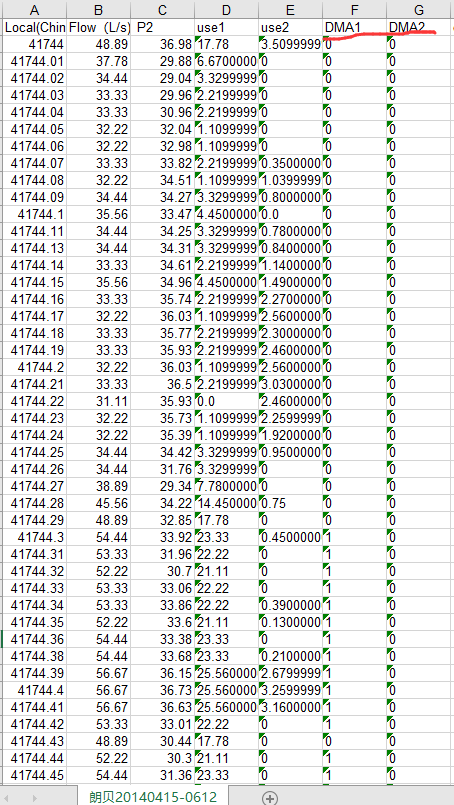
3. Abnormal pattern recognition
3.1 Extraction Characteristics
3.1.1 Calculating the Variation
def cal_change():
data = xlrd.open_workbook("result.xls") # Open excel
table = data.sheet_by_name("Lambert 20140 415-0612") # Read sheet
from xlutils.copy import copy
wb = xlutils.copy.copy(data)
sheet = wb.get_sheet(0)
sheet.write(0, 7, 'change1')
sheet.write(0, 8, 'change2')
oldRow = table.row_values(1)
for i in range(2, table.nrows):
row = table.row_values(i) # Line data in an array
sheet.write(i, 7, abs(float(row[3]) - float(oldRow[3])))
sheet.write(i, 8, abs(float(row[4]) - float(oldRow[4])))
oldRow = row
wb.save("result.xls")
Append the calculated amount of change to the back of the document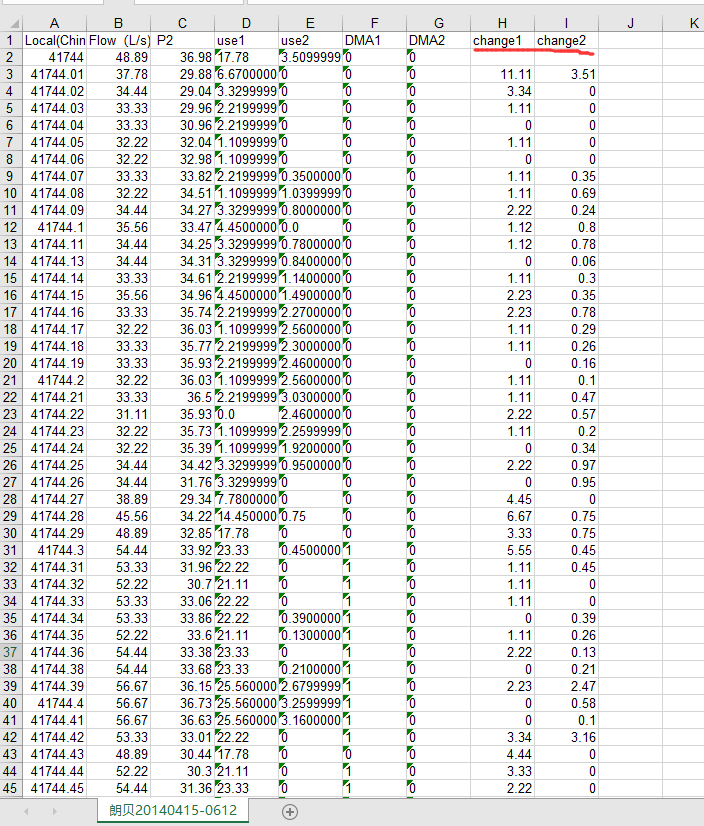
3.1.2 Construction Matrix
Constructing different matrices for clustering after different patterns
def get_change():
data = xlrd.open_workbook("result.xls") # Open excel
table = data.sheet_by_name("Lambert 20140 415-0612") # Read sheet
DMA10 = []
DMA11 = []
DMA20 = []
DMA21 = []
DMA22 = []
oldRow = table.row_values(1)
for i in range(2, table.nrows):
row = table.row_values(i) # Line data in an array
item = [float(row[3]), abs(float(row[3]) - float(oldRow[3]))]
if int(row[5]) == 0:
DMA10.append(item)
else:
DMA11.append(item)
item = [float(row[4]), abs(float(row[4]) - float(oldRow[4]))]
if int(row[6]) == 0:
DMA20.append(item)
elif int(row[6]) == 1:
DMA21.append(item)
else:
DMA22.append(item)
oldRow = row
return np.array(DMA10), np.array(DMA11),np.array(DMA20), np.array(DMA21), np.array(DMA22)
3.2DBSCAN Clustering
After clustering, the distribution map of points is drawn according to the label value.
def find_by_DBSCAN(X, eps, min_samples, name):
y_pred = DBSCAN(eps=eps, min_samples=min_samples).fit_predict(X)
plt.scatter(X[:, 0], X[:, 1], c=y_pred)
# plt.show()
plt.savefig(name)
Remember to adjust the parameters for different types. I'll give you the case of mode 0 of DMA1 with eps=10 and min_samples=10 (corresponding to mode 1 in the original text).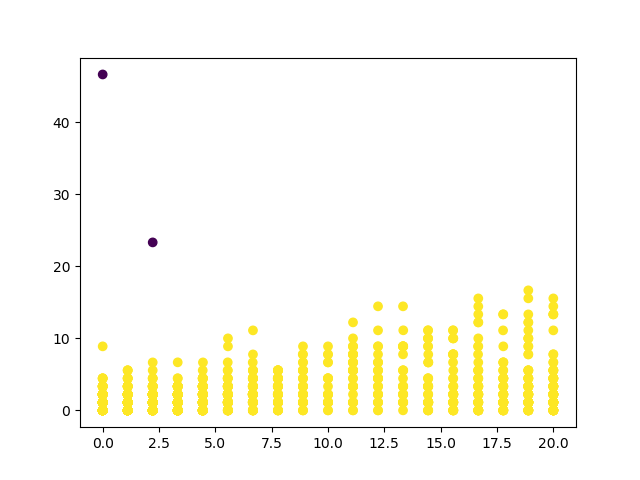
3.3 Print outliers
Call after clustering
def print_exception(lables, name):
if name == "DMA10":
columnIndex = 5
typeValue = 0
elif name == "DMA11":
columnIndex = 5
typeValue = 1
elif name == "DMA20":
columnIndex = 6
typeValue = 0
elif name == "DMA21":
columnIndex = 6
typeValue = 1
elif name == "DMA22":
columnIndex = 6
typeValue = 2
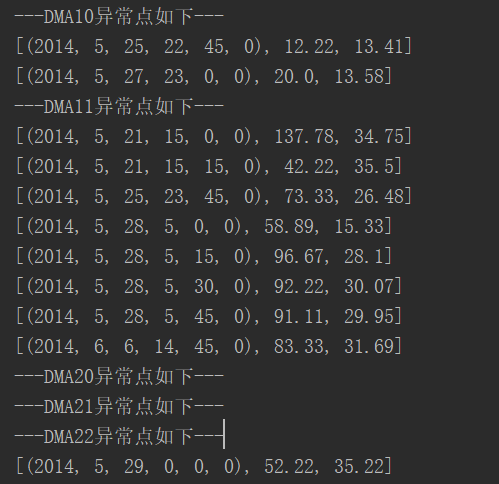
print("---"+name+"The outliers are as follows---")
data = xlrd.open_workbook("result.xls") # Open excel
table = data.sheet_by_name("Lambert 20140 415-0612") # Read sheet
count = 0
for i in range(2, table.nrows):
row = table.row_values(i) # Line data in an array
if int(row[columnIndex]) == int(typeValue):
count = count + 1
if int(lables[count - 1]) != 0:
date = xldate_as_tuple(row[0], 0)
print([date, row[1], row[2]])
It can be seen that the results are not very different from those given in the paper. It should be noted that my 0 and 1 are exactly opposite to the model 0 and 1 of the paper.